Document
... updated algorithms, how can we easily rerun analyses? What privacy software do we need and could leverage? 2. Will SageCommons need to be ‘replicable’ at other sites to support privacy - e.g. Pharma and Biotech who do not want their use of the models to be potentially snooped on the ‘net? ...
... updated algorithms, how can we easily rerun analyses? What privacy software do we need and could leverage? 2. Will SageCommons need to be ‘replicable’ at other sites to support privacy - e.g. Pharma and Biotech who do not want their use of the models to be potentially snooped on the ‘net? ...
Variable and Feature Selection in Machine Learning (Review
... variation (e.g. PCA etc) • Problem is that we are no longer dealing with one feature at a time but rather a linear or possibly more complicated combination of all features. It may be good enough for a black box but how does one build a diagnostic chip on a “supergene”? (even though we don’t want to ...
... variation (e.g. PCA etc) • Problem is that we are no longer dealing with one feature at a time but rather a linear or possibly more complicated combination of all features. It may be good enough for a black box but how does one build a diagnostic chip on a “supergene”? (even though we don’t want to ...
VanBUG_quackenbush
... Unless a reviewer has the courage to give you unqualified praise, I say ignore the bastard. ...
... Unless a reviewer has the courage to give you unqualified praise, I say ignore the bastard. ...
2.2 Distance Measures for Binary Attributes
... Given two p-dimensional instances, xi = (xi1; xi2; : : : ; xip) and xj = (xj1; xj2; : : : ; xjp), The distance between the two data instances can be calculated using the Minkowski metric (Han and Kamber, 2001): d(xi; xj) = (jxi1 ¡ xj1jg + jxi2 ¡ xj2jg + : : : + jxip ¡ xjpjg)1=g The commonly used Euc ...
... Given two p-dimensional instances, xi = (xi1; xi2; : : : ; xip) and xj = (xj1; xj2; : : : ; xjp), The distance between the two data instances can be calculated using the Minkowski metric (Han and Kamber, 2001): d(xi; xj) = (jxi1 ¡ xj1jg + jxi2 ¡ xj2jg + : : : + jxip ¡ xjpjg)1=g The commonly used Euc ...
Hierarchical Stability Based Model Selection for Data Clustering
... The correct answer of K for a given data is unknown So we need a better way to find this K and also the positions of the K centers This can be intuitively called model selection for clustering algorithms. Existing model selection method: ● Bayesian Information Criterion ● Gap statistics ● Projection ...
... The correct answer of K for a given data is unknown So we need a better way to find this K and also the positions of the K centers This can be intuitively called model selection for clustering algorithms. Existing model selection method: ● Bayesian Information Criterion ● Gap statistics ● Projection ...
A hierarchical unsupervised growing neural network for
... robust and accurate approach to the clustering of big amounts of noisy data. Neural networks have a series of properties that make them suitable for the analysis of gene expression patterns. They can deal with real-world data sets containing noisy, illdefined items with irrelevant variables and outl ...
... robust and accurate approach to the clustering of big amounts of noisy data. Neural networks have a series of properties that make them suitable for the analysis of gene expression patterns. They can deal with real-world data sets containing noisy, illdefined items with irrelevant variables and outl ...
Projected clustering
... ! The clustering process is based on the K-means algorithm ! K-means partitions a data into a number of clusters, each of which is represented by a center. ...
... ! The clustering process is based on the K-means algorithm ! K-means partitions a data into a number of clusters, each of which is represented by a center. ...
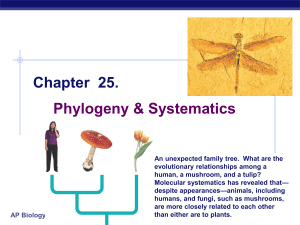
Ch_25 Phylogeny and Systematics
... genomes of different organisms, we find… humans & mice have 99% of their genes in ...
... genomes of different organisms, we find… humans & mice have 99% of their genes in ...
Lecture 8: Advanced Clustering
... ◦ Similarity measurement and clustering methods for graph and networks Clustering with Constraints ◦ Cluster analysis under different kinds of constraints, e.g., that raised from background knowledge or spatial distribution of the objects ...
... ◦ Similarity measurement and clustering methods for graph and networks Clustering with Constraints ◦ Cluster analysis under different kinds of constraints, e.g., that raised from background knowledge or spatial distribution of the objects ...
Slides Here
... Analysis of the full Forest of Life in comparison to NUTs shows that: • a considerable fraction of FOL trees are very similar to NUTs: average FOL-NUTs similarity is dramatically above the random level • unlike NUTs, topologies of the FOL trees show distinct clustering largely determined by the phyl ...
... Analysis of the full Forest of Life in comparison to NUTs shows that: • a considerable fraction of FOL trees are very similar to NUTs: average FOL-NUTs similarity is dramatically above the random level • unlike NUTs, topologies of the FOL trees show distinct clustering largely determined by the phyl ...
Phylogenetic Relationships Among Ascomycetes: Evidence from an
... PAUP, version 3.1.1 (Swofford 1993), with both equalweights parsimony and a weighted step matrix based on the JTT matrix (Felsenstein 1981; Jones, Taylor, and Thornton 1992). The heuristic search using the randomaddition-of-taxon option was performed with 100 replicates to increase the chance of fin ...
... PAUP, version 3.1.1 (Swofford 1993), with both equalweights parsimony and a weighted step matrix based on the JTT matrix (Felsenstein 1981; Jones, Taylor, and Thornton 1992). The heuristic search using the randomaddition-of-taxon option was performed with 100 replicates to increase the chance of fin ...
On the optimization of classes for the assignment of unidentified
... must take an empirical, data-driven, operational approach49. Phylogenetic methods that are based on the analysis of macromolecular sequences50,51 are bound up so intimately with the questions of evolution that they do not seem suitable for our purposes. Indeed, the biggest (and effectively insuperab ...
... must take an empirical, data-driven, operational approach49. Phylogenetic methods that are based on the analysis of macromolecular sequences50,51 are bound up so intimately with the questions of evolution that they do not seem suitable for our purposes. Indeed, the biggest (and effectively insuperab ...
compEpiTools - Bioconductor
... (identification of ’direct’ enhancers). This does not apply if those TSS belong to isoforms of the same gene. This method returns: (i) a set of reference regions without any interacting direct enhancers, (ii) a set of enhancers sites having putative taget regions, and (iii) those of putative target ...
... (identification of ’direct’ enhancers). This does not apply if those TSS belong to isoforms of the same gene. This method returns: (i) a set of reference regions without any interacting direct enhancers, (ii) a set of enhancers sites having putative taget regions, and (iii) those of putative target ...
Classification, subtype discovery, and prediction of outcome in
... • A total of 12 EPs, some important ones of them never discovered by C4.5. • Examples: {Humi <=80, windy = false} -> Play (5:0). • A total of 5 rules in the decision tree induced by C4.5. • C4.5 missed many important rules. ...
... • A total of 12 EPs, some important ones of them never discovered by C4.5. • Examples: {Humi <=80, windy = false} -> Play (5:0). • A total of 5 rules in the decision tree induced by C4.5. • C4.5 missed many important rules. ...
A computational platform for whole genome association analysis
... Test for correlation between unlinked loci Test for difference in correlation between loci, in cases and controls ...
... Test for correlation between unlinked loci Test for difference in correlation between loci, in cases and controls ...
Package `NAPPA`
... Enables the processing and normalisation of the mRNA data output from the Nanostring nCounter software. Performs an adjustment based on the observed field of view for each lane. Performs a background correction using the truncated Poisson distribution adjustment. Performs a positive control normalis ...
... Enables the processing and normalisation of the mRNA data output from the Nanostring nCounter software. Performs an adjustment based on the observed field of view for each lane. Performs a background correction using the truncated Poisson distribution adjustment. Performs a positive control normalis ...
Package `TSGSIS`
... for detection of whole-genome SNP effects and SNP-SNP interactions, as described in Fang et al. (2017, under review). The proposed TSGSIS is developed to study interactions that may not have marginal effects. ...
... for detection of whole-genome SNP effects and SNP-SNP interactions, as described in Fang et al. (2017, under review). The proposed TSGSIS is developed to study interactions that may not have marginal effects. ...
In recent year there have been rapid progress made in mapping the
... methods of analysis (a selection is provided in the reference section). These methods fall into two main classes: (i) methods that compare the groups gene-by-gene and make corrections to the p-values provided by each test; and (ii) methods that identify differentiably expressed genes by modeling the ...
... methods of analysis (a selection is provided in the reference section). These methods fall into two main classes: (i) methods that compare the groups gene-by-gene and make corrections to the p-values provided by each test; and (ii) methods that identify differentiably expressed genes by modeling the ...
Module Discovery in Gene Expression Data Using Closed Itemset
... conditions. The data used to search for expression modules typically is data from several microarray chip measurements, labeled by the experimental condition the sample was subjected to before performing the measurement. In recent years, several biclustering methods have been suggested to discover m ...
... conditions. The data used to search for expression modules typically is data from several microarray chip measurements, labeled by the experimental condition the sample was subjected to before performing the measurement. In recent years, several biclustering methods have been suggested to discover m ...
GENOTYPE-PHENOTYPE CORRELATION USING
... Biological science has undergone a revolution in the past few decades. The successes of molecular and structural biology, biochemistry, and genetics have yielded large amounts of data that are increasingly quantitative in nature. This quantitative analysis of this data has attracted the use of techn ...
... Biological science has undergone a revolution in the past few decades. The successes of molecular and structural biology, biochemistry, and genetics have yielded large amounts of data that are increasingly quantitative in nature. This quantitative analysis of this data has attracted the use of techn ...
Chapter 10 Neural Networks
... • During the learning phase, training data is used to modify the connection weights between pairs of nodes so as to obtain a best result for the output node (s). ...
... • During the learning phase, training data is used to modify the connection weights between pairs of nodes so as to obtain a best result for the output node (s). ...
An Approach to Solve Winner Determination in Combinatorial
... algorithms are not only inadequate but also infeasible as instances become larger [5]. In real-time applications, certain domains may require approximate solutions within an allowable processing time. Sometimes, it is unnecessary to expense a lot to better improve the quality of the solution. For th ...
... algorithms are not only inadequate but also infeasible as instances become larger [5]. In real-time applications, certain domains may require approximate solutions within an allowable processing time. Sometimes, it is unnecessary to expense a lot to better improve the quality of the solution. For th ...
HTSanalyzeR - Florian Markowetz
... Parameters and report. Each of these analysis methods depends on several input parameters. While every one of them can be changed in the package, HTSanalyzeR also implements a standard analysis option using default parameters that we have found to work well in many applications. Results are presente ...
... Parameters and report. Each of these analysis methods depends on several input parameters. While every one of them can be changed in the package, HTSanalyzeR also implements a standard analysis option using default parameters that we have found to work well in many applications. Results are presente ...
A DNA-sequence based phylogeny for triculine snails (Gastropoda
... test of Xia et al. (2002) as found in the DAMBE software package of Xia (1999), which provides a statistical test for saturation. The test was chosen because it was thought more likely to detect saturation in the present data, where several closely related species are compared, than say randomizatio ...
... test of Xia et al. (2002) as found in the DAMBE software package of Xia (1999), which provides a statistical test for saturation. The test was chosen because it was thought more likely to detect saturation in the present data, where several closely related species are compared, than say randomizatio ...
GenomicsResourcesForEmergingModelOrganismsPoster
... diverse contexts, from genome annotation projects within individual labs to major model organism databases. ...
... diverse contexts, from genome annotation projects within individual labs to major model organism databases. ...