Team Application Activity #3: Statistical Analysis of Microbial
... To calculate alpha diversity, QIIME must first generate alpha rarefaction tables (in biom format). As you know from your readings, rarefaction data will not only provide information regarding the amount of diversity present within each sample, but will also help you determine if you have sampled at ...
... To calculate alpha diversity, QIIME must first generate alpha rarefaction tables (in biom format). As you know from your readings, rarefaction data will not only provide information regarding the amount of diversity present within each sample, but will also help you determine if you have sampled at ...
Comparing samples—part II
... corresponding to an effect will have more P values close to 0 (Fig. 3a). In a real-world experiment we do not know which comparisons truly correspond to an effect, so all we see is the aggregate distribution, shown as the third histogram in Figure 3a. If the effect rate is low, most of our P values ...
... corresponding to an effect will have more P values close to 0 (Fig. 3a). In a real-world experiment we do not know which comparisons truly correspond to an effect, so all we see is the aggregate distribution, shown as the third histogram in Figure 3a. If the effect rate is low, most of our P values ...
Archaeal phylogenomics provides evidence in support of a
... a version of MRBAYES v. 3.1.1 to allow us to constrain branching order while allowing branch lengths to vary. Since topology is constrained, this approach allows us to place the intersection at any position in the archaeal tree and evaluate the overall likelihood of that tree once the other paramete ...
... a version of MRBAYES v. 3.1.1 to allow us to constrain branching order while allowing branch lengths to vary. Since topology is constrained, this approach allows us to place the intersection at any position in the archaeal tree and evaluate the overall likelihood of that tree once the other paramete ...
A powerful test of independent assortment that determines
... consumes a larger fraction of the total time needed to compute the adjusted P-value. In fact, when the analysis programs make use of all of the available data (for example, EAGLET (Stewart et al., 2010; Stewart et al., 2011, 2013; Kambhampati et al., 2013) and MORGAN (Thompson, 1994; Heath et al., 1 ...
... consumes a larger fraction of the total time needed to compute the adjusted P-value. In fact, when the analysis programs make use of all of the available data (for example, EAGLET (Stewart et al., 2010; Stewart et al., 2011, 2013; Kambhampati et al., 2013) and MORGAN (Thompson, 1994; Heath et al., 1 ...
pplacer: linear time maximum-likelihood and Bayesian phylogenetic
... tool for the evolutionary analysis of sequence data. It has well-developed statistical foundations for inference [14,15], tests for uncertainty estimation [16], and sophisticated evolutionary models [17,18]. In contrast to distance-based methods, likelihood-based methods can use both low and high va ...
... tool for the evolutionary analysis of sequence data. It has well-developed statistical foundations for inference [14,15], tests for uncertainty estimation [16], and sophisticated evolutionary models [17,18]. In contrast to distance-based methods, likelihood-based methods can use both low and high va ...
A New Method for Estimating the Risk Ratio in Studies Using Case
... of Khoury's method or Flanders and Khoury's method and that it is slightly larger than that of the maximum likelihood-based method of Schaid and Sommer. Despite the slightly large variance of the new estimator compared with that of the maximum likelihood-based method, the simplicity of the new estim ...
... of Khoury's method or Flanders and Khoury's method and that it is slightly larger than that of the maximum likelihood-based method of Schaid and Sommer. Despite the slightly large variance of the new estimator compared with that of the maximum likelihood-based method, the simplicity of the new estim ...
Bioinformatics Dr. Víctor Treviño Pabellón Tec
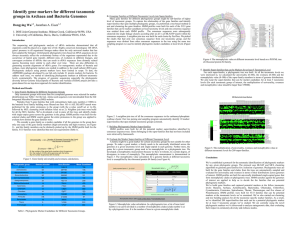
... more (multiple sequence alignment) sequences by searching for similar patterns that are in the same order in the sequences ...
... more (multiple sequence alignment) sequences by searching for similar patterns that are in the same order in the sequences ...
Full-text PDF
... results if K is set to any value larger than 4. They themselves use the setting K = 7 in their experiments against the Daly et al.'s data [6]. So we also set K to 7 in our experiments in section 3. Once the model has been trained, we can estimate haplotypes from genotypes. Moreover we can obtain mul ...
... results if K is set to any value larger than 4. They themselves use the setting K = 7 in their experiments against the Daly et al.'s data [6]. So we also set K to 7 in our experiments in section 3. Once the model has been trained, we can estimate haplotypes from genotypes. Moreover we can obtain mul ...
Full-text PDF
... Figure 1: In these GenBank Release 110 entries for two different organisms, the strategies used for storing ORF ID (bold type) and gene name (underlined) information are inconsistent. • In the transformation approach, users need to know some details about the original data formats to be transformed, ...
... Figure 1: In these GenBank Release 110 entries for two different organisms, the strategies used for storing ORF ID (bold type) and gene name (underlined) information are inconsistent. • In the transformation approach, users need to know some details about the original data formats to be transformed, ...
Identification of Short Motifs for Comparing Biological Sequences
... from the fact that many of the compression algorithms could be implemented in a linear time complexity. Compressionbased techniques also showed very good quality with the results, especially those techniques that are dictionarybased. The two major techniques for compression are Lempel-Ziv complexity ...
... from the fact that many of the compression algorithms could be implemented in a linear time complexity. Compressionbased techniques also showed very good quality with the results, especially those techniques that are dictionarybased. The two major techniques for compression are Lempel-Ziv complexity ...
We need an optimality criterion to choose a best estimate (tree
... least amount of change along its branches to produce the data. ...
... least amount of change along its branches to produce the data. ...
Evaluation of Nyholt`s Procedure for Multiple Testing Correction
... Dudbridge and Koeleman (2004) investigated whether the assumption underlying Nyholt’s method, that there really is an ‘effective’ number of independent tests, is true. When b independent tests are carried out, the minimum p-value has a Beta(1, b) distribution. Using data on chromosomes 18 and 21 fro ...
... Dudbridge and Koeleman (2004) investigated whether the assumption underlying Nyholt’s method, that there really is an ‘effective’ number of independent tests, is true. When b independent tests are carried out, the minimum p-value has a Beta(1, b) distribution. Using data on chromosomes 18 and 21 fro ...
Metabolomics - Horticultural Sciences at University of Florida
... Thus, in principle, the function of an unknown gene can be determined by comparing the metabolic profile of a mutant in that gene with a library of such profiles generated by deleting individual genes of known function. Caution: This approach may not be so useful for dissecting metabolic responses t ...
... Thus, in principle, the function of an unknown gene can be determined by comparing the metabolic profile of a mutant in that gene with a library of such profiles generated by deleting individual genes of known function. Caution: This approach may not be so useful for dissecting metabolic responses t ...
Combining Machine Learning and Homology-Based
... of conservation against mutations to 20 different amino acids, including itself. A matrix consisting of such vector representations for all the residues in a given sequence is called the PSSM. When a residue is conserved through cycles of PSI-BLAST, it is likely to be due to a purpose (i.e. biologic ...
... of conservation against mutations to 20 different amino acids, including itself. A matrix consisting of such vector representations for all the residues in a given sequence is called the PSSM. When a residue is conserved through cycles of PSI-BLAST, it is likely to be due to a purpose (i.e. biologic ...
a review of methods for encoding neural network topologies in
... 2) Koza node-based encoding Another possibility of node-based encoding is to use genetic programming. Since GP is usually applied to evolve program trees in LISP language, the network in this method is represented as a tree, where the root is the output processing element (neuron) and the leaves rep ...
... 2) Koza node-based encoding Another possibility of node-based encoding is to use genetic programming. Since GP is usually applied to evolve program trees in LISP language, the network in this method is represented as a tree, where the root is the output processing element (neuron) and the leaves rep ...
User Manual of ClusterProject
... The column of Rep is indispensable whether the experiment has replication or not. If there is no replication, all values of this column are set to one. It can have additional factors in the input file such as dye, treatment or array et al. This is tab-delimited text file. Mixed model approaches are ...
... The column of Rep is indispensable whether the experiment has replication or not. If there is no replication, all values of this column are set to one. It can have additional factors in the input file such as dye, treatment or array et al. This is tab-delimited text file. Mixed model approaches are ...
Discovering biclusters in gene expression data based on high
... It should be pointed out that some symbolic, coherent evolution or numerical biclusters, such as those produced by cMonkey [9], SAMBA [10] and some statistical criteria, cannot be classified as additive or multiplicative patterns directly. For example, in cMonkey, additional information besides the ...
... It should be pointed out that some symbolic, coherent evolution or numerical biclusters, such as those produced by cMonkey [9], SAMBA [10] and some statistical criteria, cannot be classified as additive or multiplicative patterns directly. For example, in cMonkey, additional information besides the ...
A microarray gene expression data classification using hybrid back
... The effects of the parameters of parallel GAs on the quality of their search and on their efficiency are not well understood. This insufficient knowledge limits our ability to design fast and accurate parallel GAs that reach the desired solutions in the shortest time possible. The goal of this disse ...
... The effects of the parameters of parallel GAs on the quality of their search and on their efficiency are not well understood. This insufficient knowledge limits our ability to design fast and accurate parallel GAs that reach the desired solutions in the shortest time possible. The goal of this disse ...
DYNAMIC BLOCK ALLOCATION FOR BIOLOGICAL SEQUENCES
... managed to find a number a, which provides t variable a value larger than three. The next step consists in finding the optimal length for data blocks in accordance with t variable. Variable r is a multiple of a, thus the difference L – t will ensure a number divisible at least by three integers. The ...
... managed to find a number a, which provides t variable a value larger than three. The next step consists in finding the optimal length for data blocks in accordance with t variable. Variable r is a multiple of a, thus the difference L – t will ensure a number divisible at least by three integers. The ...
The development of restriction analysis and PCR
... The selection of PCR primers was dictated by similar considerations as the selection of enzymes for the restriction analysis. Primer 1 is complementary to the sense (+) strand such that the 3’ end is towards (but short of) the BamH1 and EcoR1 restriction sites. Thus, it is the forward primer. Primer ...
... The selection of PCR primers was dictated by similar considerations as the selection of enzymes for the restriction analysis. Primer 1 is complementary to the sense (+) strand such that the 3’ end is towards (but short of) the BamH1 and EcoR1 restriction sites. Thus, it is the forward primer. Primer ...
Revealing the demographic histories of species
... estimating demographic history from gene sequence data using statistical models that were originally designed for the analysis of survival data23–25. The data used are divergence times among a group of sequences as estimated from a phylogenetic tree. The number of lineages within a reconstructed phy ...
... estimating demographic history from gene sequence data using statistical models that were originally designed for the analysis of survival data23–25. The data used are divergence times among a group of sequences as estimated from a phylogenetic tree. The number of lineages within a reconstructed phy ...
A Step-by-Step Tutorial: Divergence Time Estimation with
... each partition must have the same number of species present in the corresponding in.BV partitions. For example, you could be analyzing tRNA genes from 20 species and amino acid sequences from 30, so the alignment file would need to have 20 and 30 species in each partition respectively. The master tr ...
... each partition must have the same number of species present in the corresponding in.BV partitions. For example, you could be analyzing tRNA genes from 20 species and amino acid sequences from 30, so the alignment file would need to have 20 and 30 species in each partition respectively. The master tr ...
Document
... • There are reference databases based on structural information: e.g. BAliBASE and HOMSTRAD • Conflicting standards of truth – evolution – structure – function ...
... • There are reference databases based on structural information: e.g. BAliBASE and HOMSTRAD • Conflicting standards of truth – evolution – structure – function ...
Keystone2011poster
... The sequencing and phylogenetic analysis of rRNA molecules demonstrated that all organisms could be placed on a single tree of life. Highly conserved, homologous 16S rRNA genes' presence in all organismal lineages makes them the only universal marker that has been adopted by biologist. Unfortunately ...
... The sequencing and phylogenetic analysis of rRNA molecules demonstrated that all organisms could be placed on a single tree of life. Highly conserved, homologous 16S rRNA genes' presence in all organismal lineages makes them the only universal marker that has been adopted by biologist. Unfortunately ...
XML MINING USING GENETIC ALGORITHM
... for data exchange over the web. Mining XML data from the web is becoming increasingly important as well. In general frequent itemsets are generated from large data sets by applying association rule mining algorithms like Apriori, Partition, Pincer-Search, Incremental, and Border algorithm etc., whic ...
... for data exchange over the web. Mining XML data from the web is becoming increasingly important as well. In general frequent itemsets are generated from large data sets by applying association rule mining algorithms like Apriori, Partition, Pincer-Search, Incremental, and Border algorithm etc., whic ...