Quiz 3 Key - UW Canvas
... 3. (5 pts) a. (2 pts) Imagine you have a wild-type (normal) E. coli cell. It is being grown in glycerol, with neither glucose nor lactose present. Circle all proteins that will be bound to the DNA of the Lac Operon or its regulatory regions. CAP protein (with cAMP) ...
... 3. (5 pts) a. (2 pts) Imagine you have a wild-type (normal) E. coli cell. It is being grown in glycerol, with neither glucose nor lactose present. Circle all proteins that will be bound to the DNA of the Lac Operon or its regulatory regions. CAP protein (with cAMP) ...
Slides
... Quantitatively characterize interactions of network elements; Predict the function of genes in biological networks. ...
... Quantitatively characterize interactions of network elements; Predict the function of genes in biological networks. ...
Molecular genetics of gene expression
... What are some biological roles of transcription factors? • Basal transcription regulation – general transcription factors • Development • Response to intercellular signals • Response to environment • Cell cycle control ...
... What are some biological roles of transcription factors? • Basal transcription regulation – general transcription factors • Development • Response to intercellular signals • Response to environment • Cell cycle control ...
S1.Describe how the tight packing of chromatin in a closed
... may prevent transcription factors and/or RNA polymerase from binding to the major groove of the DNA. Second, it may prevent RNA polymerase from forming an open complex, which is necessary to begin transcription. Third, it could prevent looping in the DNA, which may be necessary to activate transcrip ...
... may prevent transcription factors and/or RNA polymerase from binding to the major groove of the DNA. Second, it may prevent RNA polymerase from forming an open complex, which is necessary to begin transcription. Third, it could prevent looping in the DNA, which may be necessary to activate transcrip ...
Document
... may prevent transcription factors and/or RNA polymerase from binding to the major groove of the DNA. Second, it may prevent RNA polymerase from forming an open complex, which is necessary to begin transcription. Third, it could prevent looping in the DNA, which may be necessary to activate transcrip ...
... may prevent transcription factors and/or RNA polymerase from binding to the major groove of the DNA. Second, it may prevent RNA polymerase from forming an open complex, which is necessary to begin transcription. Third, it could prevent looping in the DNA, which may be necessary to activate transcrip ...
Chapter 4- Genes and development
... • Getting the DNA (a gene) into a cell– ______________ – __________________ (mix DNA with cells) – Retrovirus _____________ (infect a cell) • P element in Drosophila- a transposable element that allows a gene to be inserted into specific positions in ...
... • Getting the DNA (a gene) into a cell– ______________ – __________________ (mix DNA with cells) – Retrovirus _____________ (infect a cell) • P element in Drosophila- a transposable element that allows a gene to be inserted into specific positions in ...
Topic 3 The Chemistry of Life - wfs
... 4. The genetic code is actually composed of triplets of bases called codons. The codons are present on the RNA formed during translation. Therefore, codons do not contain thymine. 5. The RNA formed during transcription is called messenger or mRNA. This mRNA carries the genetic code out of the nucleu ...
... 4. The genetic code is actually composed of triplets of bases called codons. The codons are present on the RNA formed during translation. Therefore, codons do not contain thymine. 5. The RNA formed during transcription is called messenger or mRNA. This mRNA carries the genetic code out of the nucleu ...
activators
... a technique used to determine if enhancer action requires DNA looping • Used to test whether two remote DNA regions, such as an enhancer and a promoter, are brought together ...
... a technique used to determine if enhancer action requires DNA looping • Used to test whether two remote DNA regions, such as an enhancer and a promoter, are brought together ...
Transcription Eukary 2
... • RNA Pol I transcribes genes for the large rRNA precursor • There are hundreds of similar copies of this gene in each genome • RNA Pol I promoters (called class I) have two components: • Upstream control element: –156 to -107 • Core element: –45 to +20 • Two different transcription factors bind the ...
... • RNA Pol I transcribes genes for the large rRNA precursor • There are hundreds of similar copies of this gene in each genome • RNA Pol I promoters (called class I) have two components: • Upstream control element: –156 to -107 • Core element: –45 to +20 • Two different transcription factors bind the ...
Summary - EUR RePub
... RNA polymerase II mainly transcribes protein-encoding genes, and the transcriptional activity of many of those genes is tightly regulated. Two cooperating components are key in regulating RNAP II transcription. One component acts in cis and requires an element that is located on the same DNA molecul ...
... RNA polymerase II mainly transcribes protein-encoding genes, and the transcriptional activity of many of those genes is tightly regulated. Two cooperating components are key in regulating RNAP II transcription. One component acts in cis and requires an element that is located on the same DNA molecul ...
Gene Regulation
... transcription and translation. Best time to control gene expression is at transcription, before a mRNA is produced. Regulation in Prokaryotes Two types of regulation in prokaryotes: 1. Inducible - Turn on production of a protein only when it is needed. 2. Repressible - turn off production of a prote ...
... transcription and translation. Best time to control gene expression is at transcription, before a mRNA is produced. Regulation in Prokaryotes Two types of regulation in prokaryotes: 1. Inducible - Turn on production of a protein only when it is needed. 2. Repressible - turn off production of a prote ...
Chapter 24: Promoters and Enhancers
... • Demethylation at the 5’ end of the gene and the promoter region is necessary for transcription. • CpG islands surround the promoters of constitutively expressed genes where they are unmethylated. • They are also found at the promoters of some tissue-regulated genes. • There are ~29,000 CpG islands ...
... • Demethylation at the 5’ end of the gene and the promoter region is necessary for transcription. • CpG islands surround the promoters of constitutively expressed genes where they are unmethylated. • They are also found at the promoters of some tissue-regulated genes. • There are ~29,000 CpG islands ...
Transcription Factors (from Wray et al Mol Biol Evol 20:1377)
... duplicated regions in the chromosomes (Chr I to V) are indicated by boxes of the same color (adapted from TIGR). The total number of bHLH genes per chromosome is indicated at the top of each chromosome in parentheses. The scale is in megabases (Mb) and is adapted from the scale available on the TIGR ...
... duplicated regions in the chromosomes (Chr I to V) are indicated by boxes of the same color (adapted from TIGR). The total number of bHLH genes per chromosome is indicated at the top of each chromosome in parentheses. The scale is in megabases (Mb) and is adapted from the scale available on the TIGR ...
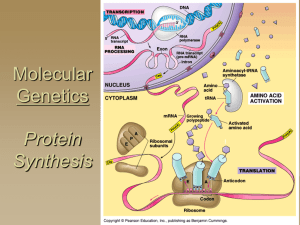
Ch. 17: From Gene to Protein
... Polypeptide marked by signal peptide (20a.a at leading end, 5’ cap modified) Recognized by Signal Recognition Particle (SRP) ...
... Polypeptide marked by signal peptide (20a.a at leading end, 5’ cap modified) Recognized by Signal Recognition Particle (SRP) ...
Eukaryotic vs. Prokaryotic genes Eukaryotic Genes
... Gene Duplication and Evolution Gene duplication events are very important in the evolution of new functions for old genes. Genes that are similar to other genes in the same genome are called paralogs. Duplications allow mutation without loss of old ...
... Gene Duplication and Evolution Gene duplication events are very important in the evolution of new functions for old genes. Genes that are similar to other genes in the same genome are called paralogs. Duplications allow mutation without loss of old ...
BIOL 112 – Principles of Zoology
... multicelled organism involves a series of “genetic switches” that regulate a cascade of developmental events ...
... multicelled organism involves a series of “genetic switches” that regulate a cascade of developmental events ...
Slide 1
... In the previous lecture you learnt that cAMP is produced as a result of activation of adenylate cyclase by G protein which in turn had been acivated by hormone which can not pass the hydrophobic cell membrane. This cAMP in turn acivates a cAMP depedent protein kinase (PKA) which modulates multiple a ...
... In the previous lecture you learnt that cAMP is produced as a result of activation of adenylate cyclase by G protein which in turn had been acivated by hormone which can not pass the hydrophobic cell membrane. This cAMP in turn acivates a cAMP depedent protein kinase (PKA) which modulates multiple a ...
Hebbian Learning by a Simple Gene Circuit
... classically conditioned therefore it must contain an intracellular circuit capable of associative learning. Hebbian learning was proposed as a mechanism for associative learning in neurons whereby synaptic weights would increase as a product of the correlated firing of pre and post synaptic neurons. ...
... classically conditioned therefore it must contain an intracellular circuit capable of associative learning. Hebbian learning was proposed as a mechanism for associative learning in neurons whereby synaptic weights would increase as a product of the correlated firing of pre and post synaptic neurons. ...
Brooker Chapter 11
... * often found nearby (-50 to -100) but can also be found great distances away in either direction ...
... * often found nearby (-50 to -100) but can also be found great distances away in either direction ...
Transcription start sites
... DNase I hypersensitive sites • In total, about 3 million DNase I hypersensitive sites in the genome, covering about 15% (versus about 40,000 genes covering about 4%) • Transcriptional start sites are regions of DNase I hypersensitivity, as expected • Most DNase I hypersensitive sites are not associ ...
... DNase I hypersensitive sites • In total, about 3 million DNase I hypersensitive sites in the genome, covering about 15% (versus about 40,000 genes covering about 4%) • Transcriptional start sites are regions of DNase I hypersensitivity, as expected • Most DNase I hypersensitive sites are not associ ...
DNA binding shifts the redox potential of the transcription factor SoxR
... contains a [2Fe-2S] cluster and is activated through oxidation. A DNA-bound potential of +200 mV versus NHE (normal hydrogen electrode) is found for SoxR isolated from Escherichia coli and Pseudomonas aeruginosa. This potential value corresponds to a dramatic shift of +490 mV versus values found in ...
... contains a [2Fe-2S] cluster and is activated through oxidation. A DNA-bound potential of +200 mV versus NHE (normal hydrogen electrode) is found for SoxR isolated from Escherichia coli and Pseudomonas aeruginosa. This potential value corresponds to a dramatic shift of +490 mV versus values found in ...
Proximal promoter
... the distal sequence upstream of the gene that may contain additional regulatory elements, often with a weaker influence than the proximal promoter – Anything further upstream (but not an enhancer or other regulatory region whose influence is positional/orientation independent) – Specific transcripti ...
... the distal sequence upstream of the gene that may contain additional regulatory elements, often with a weaker influence than the proximal promoter – Anything further upstream (but not an enhancer or other regulatory region whose influence is positional/orientation independent) – Specific transcripti ...
Transcription factor
In molecular biology and genetics, a transcription factor (sometimes called a sequence-specific DNA-binding factor) is a protein that binds to specific DNA sequences, thereby controlling the rate of transcription of genetic information from DNA to messenger RNA. Transcription factors perform this function alone or with other proteins in a complex, by promoting (as an activator), or blocking (as a repressor) the recruitment of RNA polymerase (the enzyme that performs the transcription of genetic information from DNA to RNA) to specific genes.A defining feature of transcription factors is that they contain one or more DNA-binding domains (DBDs), which attach to specific sequences of DNA adjacent to the genes that they regulate. Additional proteins such as coactivators, chromatin remodelers, histone acetylases, deacetylases, kinases, and methylases, while also playing crucial roles in gene regulation, lack DNA-binding domains, and, therefore, are not classified as transcription factors.