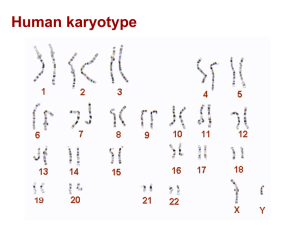
regulatory-network
... Sequence alignment is a good way to infer protein function, when two proteins do the exact same thing in two different organisms. ...
... Sequence alignment is a good way to infer protein function, when two proteins do the exact same thing in two different organisms. ...
Plant Transformation
... • nucleic acid sequences encoding easily assayed proteins • Reporter genes include -galactosidase (encoded by lacZ), -glucuronidase (encoded by uidA), chloramphenicol acetyltransferase, luciferase and green fluorescent protein (GFP) . ...
... • nucleic acid sequences encoding easily assayed proteins • Reporter genes include -galactosidase (encoded by lacZ), -glucuronidase (encoded by uidA), chloramphenicol acetyltransferase, luciferase and green fluorescent protein (GFP) . ...
Norwich_Bielski_Hulsebris_Smith_Latshaw
... Gene ontology • My gene ICL1 has been assigned the following GO categories isocitrate lyase activity (IMP, ISS), ...
... Gene ontology • My gene ICL1 has been assigned the following GO categories isocitrate lyase activity (IMP, ISS), ...
Lesson 16.1 Genes and Variation
... a) Independent assortment ____________________________________ b)Crossing over ___________________________________________ c) Random fertilization (through sexual __________________________ ___________________________________________________________ __________________________________________________ ...
... a) Independent assortment ____________________________________ b)Crossing over ___________________________________________ c) Random fertilization (through sexual __________________________ ___________________________________________________________ __________________________________________________ ...
Slide 1
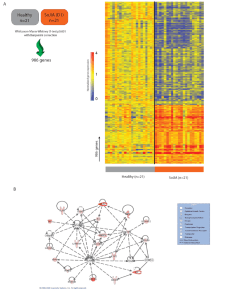
... A. Data were normalized in Beadstudio using the "average" method and imported into Genespring 7.3 (Agilent) where the expression value for each gene was normalized to the median expression value of that gene’s measurement in the healthy controls. To identify transcripts differentially expressed betw ...
... A. Data were normalized in Beadstudio using the "average" method and imported into Genespring 7.3 (Agilent) where the expression value for each gene was normalized to the median expression value of that gene’s measurement in the healthy controls. To identify transcripts differentially expressed betw ...
Gene linkage ppt
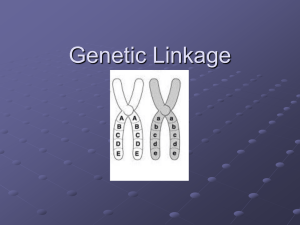
... Linked genes are pairs or groups of genes which are inherited together, carried on the same chromosome (usually close together) ...
... Linked genes are pairs or groups of genes which are inherited together, carried on the same chromosome (usually close together) ...
國立嘉義大學九十七學年度
... exons is 266 base pairs. (i) Calculate the fraction of nucleotides in a gene that will be present in mRNA? (ii) What the fraction of the chromosome that is occupied by genes? (iii) Propose a simple procedure that would help us to establish the intron/exon structure of a gene. (9%) (2) How many diffe ...
... exons is 266 base pairs. (i) Calculate the fraction of nucleotides in a gene that will be present in mRNA? (ii) What the fraction of the chromosome that is occupied by genes? (iii) Propose a simple procedure that would help us to establish the intron/exon structure of a gene. (9%) (2) How many diffe ...
Expression of yolk protein genes in liver Beekman, Johanna
... thosecells and expressed.lncreasingthe oestrogen receptor concentrationartificiatly,makesthese cells a good host for gene transfer experimentsin the study of oestrogen-regulatedgene expression.Weused the CEHcells to identify regulatory regions of the Vtg gene (Chapter3). In this chapter we confirmth ...
... thosecells and expressed.lncreasingthe oestrogen receptor concentrationartificiatly,makesthese cells a good host for gene transfer experimentsin the study of oestrogen-regulatedgene expression.Weused the CEHcells to identify regulatory regions of the Vtg gene (Chapter3). In this chapter we confirmth ...
Expression of yolk protein genes in liver Beekman, Johanna
... thosecells and expressed.lncreasingthe oestrogen receptor concentrationartificiatly,makesthese cells a good host for gene transfer experimentsin the study of oestrogen-regulatedgene expression.Weused the CEHcells to identify regulatory regions of the Vtg gene (Chapter3). In this chapter we confirmth ...
... thosecells and expressed.lncreasingthe oestrogen receptor concentrationartificiatly,makesthese cells a good host for gene transfer experimentsin the study of oestrogen-regulatedgene expression.Weused the CEHcells to identify regulatory regions of the Vtg gene (Chapter3). In this chapter we confirmth ...
Targeted knock-up of endogenous genes using a
... increase the amount of protein made by a targeted endogenous gene. This technology was first demonstrated in an elegant study by Carrieri et al (Nature 491:454). This paper describes a particular lncRNA containing a SINEB2 repeat that increases the efficiency of protein translation for a target gene ...
... increase the amount of protein made by a targeted endogenous gene. This technology was first demonstrated in an elegant study by Carrieri et al (Nature 491:454). This paper describes a particular lncRNA containing a SINEB2 repeat that increases the efficiency of protein translation for a target gene ...
GENETICS UNIT STUDY GUIDE
... and an organism’s DNA may be changed. The methods used to produce new forms of DNA are called genetic ...
... and an organism’s DNA may be changed. The methods used to produce new forms of DNA are called genetic ...
Gene Section SEPT5 (septin 5) Atlas of Genetics and Cytogenetics
... just 5'of GPIb beta (platelet membrane glycoprotein Ib beta precursor), and GPIb beta is co-expressed with hCDCRel-1; this is due to a non-consensus polyadenylation signal in 3' of hCDCRel-1. ...
... just 5'of GPIb beta (platelet membrane glycoprotein Ib beta precursor), and GPIb beta is co-expressed with hCDCRel-1; this is due to a non-consensus polyadenylation signal in 3' of hCDCRel-1. ...
AACR and other questions to be used as extra credit at end of 2150
... encodes for a protein required for blood clotting. The mutation results in no protein product being made from this gene. Each of them has one copy of this mutation. Ignore the possible effects of X inactivation. How would a biologist: A. explain how this mutation could affect the woman? B. explain h ...
... encodes for a protein required for blood clotting. The mutation results in no protein product being made from this gene. Each of them has one copy of this mutation. Ignore the possible effects of X inactivation. How would a biologist: A. explain how this mutation could affect the woman? B. explain h ...
Gene Editing - Royal Society of New Zealand
... An organism’s genetic material, or genome, is made of long molecules of DNA. These carry instructions on how to build that organism, like a manual which tells all the cells in the body how to behave. ...
... An organism’s genetic material, or genome, is made of long molecules of DNA. These carry instructions on how to build that organism, like a manual which tells all the cells in the body how to behave. ...
Document
... Representation of predicted R gene product structures and a model coupling the recognition of microbial Avr-dependent ligand and activation of plant defense. Pto can directly bind AvrPto (83, 92). The other R proteins probably bind the corresponding Avr gene products, either directly or in associati ...
... Representation of predicted R gene product structures and a model coupling the recognition of microbial Avr-dependent ligand and activation of plant defense. Pto can directly bind AvrPto (83, 92). The other R proteins probably bind the corresponding Avr gene products, either directly or in associati ...
Microarrays - TeacherWeb
... organism during different life stages • Compare gene expression of the same organism in different environments ...
... organism during different life stages • Compare gene expression of the same organism in different environments ...
What should be known about human gene nomenclature in - C-HPP
... numbers to all human protein-coding genes. As stated on their web site HGNC has "assigned unique gene symbols and names to over 33,000 human loci, of which around 19,000 are protein coding." UniProt and thus also neXtProt has tried, for the more than 1'000 genes with no official gene symbol to assig ...
... numbers to all human protein-coding genes. As stated on their web site HGNC has "assigned unique gene symbols and names to over 33,000 human loci, of which around 19,000 are protein coding." UniProt and thus also neXtProt has tried, for the more than 1'000 genes with no official gene symbol to assig ...
What is the most likely path of inheritance?
... Paula and Bernie have a child named Liz. Paula is A – and Liz is B +. What are the possible blood phenotypes for Bernie? Genotypes for all? ...
... Paula and Bernie have a child named Liz. Paula is A – and Liz is B +. What are the possible blood phenotypes for Bernie? Genotypes for all? ...
Powerpoint Presentation: Gene Transfer
... Every time the bacterium divides the plasmid is replicated too Gene expressed by the bacterium Same protein is synthesised Universal genetic code Human proteins can be produced by bacteria E.g. Humulin (Human Insulin) E.g. Human somatotropin (growth ...
... Every time the bacterium divides the plasmid is replicated too Gene expressed by the bacterium Same protein is synthesised Universal genetic code Human proteins can be produced by bacteria E.g. Humulin (Human Insulin) E.g. Human somatotropin (growth ...