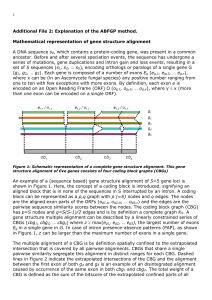
Guide for Bioinformatics Project Module 3 - SGD-Wiki
... what may be the most important residues in this domain based on those that have been unchanged throughout evolution. To do this HMM Logos provide the researcher with a quick overview of ...
... what may be the most important residues in this domain based on those that have been unchanged throughout evolution. To do this HMM Logos provide the researcher with a quick overview of ...
PPTX - Tandy Warnow
... • MetaPhyler, MetaPhlAn, and mOTU are marker-based techniques (but use different marker genes). ...
... • MetaPhyler, MetaPhlAn, and mOTU are marker-based techniques (but use different marker genes). ...
A Genetic Algorithm Approach to Solve for Multiple Solutions of
... A Genetic Algorithm, through Selection, Cross-over and Mutation operations, finds the individuals that have the best fitness values and combines them to produce individuals that offer better fitness values than their parents. This process continues until the population converges around the single indiv ...
... A Genetic Algorithm, through Selection, Cross-over and Mutation operations, finds the individuals that have the best fitness values and combines them to produce individuals that offer better fitness values than their parents. This process continues until the population converges around the single indiv ...
Quantile Regression for Large-scale Applications
... ates (Koenker & Bassett, 1978), in a manner analogous to the way in which Least-squares regression estimates the conditional mean. The Least Absolute Deviations regression (i.e., `1 regression) is a special case of quantile regression that involves computing the median of the conditional distributio ...
... ates (Koenker & Bassett, 1978), in a manner analogous to the way in which Least-squares regression estimates the conditional mean. The Least Absolute Deviations regression (i.e., `1 regression) is a special case of quantile regression that involves computing the median of the conditional distributio ...
what is alignment? - UWI St. Augustine
... Significance of local sequence alignment •In global alignment, an attempt is made to align the entire sequences, as many characters as possible. • In local alignment, stretches of sequence with the highest density of matches are given the highest priority, •generating one or more islands of matches ...
... Significance of local sequence alignment •In global alignment, an attempt is made to align the entire sequences, as many characters as possible. • In local alignment, stretches of sequence with the highest density of matches are given the highest priority, •generating one or more islands of matches ...
Halpotyping - CS, Technion
... In this discussion was presented a new method, Gibbs-Jump, for haplotype analysis, which explores the whole distribution of haplotypes conditional on the observed phenotypes. The method is very time-efficient. The result accuracy was compared to obtained by other methods (described by Sobol). ...
... In this discussion was presented a new method, Gibbs-Jump, for haplotype analysis, which explores the whole distribution of haplotypes conditional on the observed phenotypes. The method is very time-efficient. The result accuracy was compared to obtained by other methods (described by Sobol). ...
Construction of PANM Database (Protostome DB) for rapid
... A stand-alone BLAST server is available that provides a convenient and amenable platform for the analysis of molluscan sequence information especially the EST sequences generated by traditional sequencing methods. However, it is found that the server has limitations in the annotation of molluscan se ...
... A stand-alone BLAST server is available that provides a convenient and amenable platform for the analysis of molluscan sequence information especially the EST sequences generated by traditional sequencing methods. However, it is found that the server has limitations in the annotation of molluscan se ...
1 Divide and Conquer with Reduce
... to believe that any such algorithms will have to keep a cumulative “sum,” computing each output value by relying on the “sum” of the all values before it. In this lecture, we’ll see a technique that allow us to implement scan in parallel. Let’s talk about another algorithmic technique: contraction. ...
... to believe that any such algorithms will have to keep a cumulative “sum,” computing each output value by relying on the “sum” of the all values before it. In this lecture, we’ll see a technique that allow us to implement scan in parallel. Let’s talk about another algorithmic technique: contraction. ...
from Terrel Smith`s class, MS-Powerpoint slide set
... – A statement of this form is very useful in proving that algorithms terminate in a finite number of steps • If a given set is in decreasing order, meaning , and for all i, i ≥ 1, the sequence is finite because the well-ordering principle states that the set has a least element . This set can repres ...
... – A statement of this form is very useful in proving that algorithms terminate in a finite number of steps • If a given set is in decreasing order, meaning , and for all i, i ≥ 1, the sequence is finite because the well-ordering principle states that the set has a least element . This set can repres ...
IOSR Journal of Computer Engineering (IOSR-JCE)
... a kind of heuristic random search algorithm based on group difference and the optimization is finished using real encoding in continuous space. But it is incapable of “survival of the fittest” during the selection process. The mutation operation in the algorithm only aims at the newly generated indi ...
... a kind of heuristic random search algorithm based on group difference and the optimization is finished using real encoding in continuous space. But it is incapable of “survival of the fittest” during the selection process. The mutation operation in the algorithm only aims at the newly generated indi ...