Lecture 27
... Certain amino acids are more likely to be accepted than others. Distribution of amino acids in proteins is not uniform (9.5% are Leu on average and only 1.2% are Trp). Can also be affected by shifts in the sequence resulting from insertion or deletion of one or more residues within a chain. Example ...
... Certain amino acids are more likely to be accepted than others. Distribution of amino acids in proteins is not uniform (9.5% are Leu on average and only 1.2% are Trp). Can also be affected by shifts in the sequence resulting from insertion or deletion of one or more residues within a chain. Example ...
Comp. Genomics
... omitted by the model ψi • Can be computed efficiently by Felsenstein’s “pruning algorithm” (recitation 6) • Joint probability of a path in the HMM and and alignment X • Viterbi, forward-backward etc. – as usual ...
... omitted by the model ψi • Can be computed efficiently by Felsenstein’s “pruning algorithm” (recitation 6) • Joint probability of a path in the HMM and and alignment X • Viterbi, forward-backward etc. – as usual ...
Learning Algorithms for Solving MDPs References: Barto, Bradtke
... References: Barto, Bradtke and Singh (1995) “Learning to Act Using Real-Time Dynamic Programming” in Machine Learning (also on WWW) 1. Q-Learning Given an MDP problem, define the ...
... References: Barto, Bradtke and Singh (1995) “Learning to Act Using Real-Time Dynamic Programming” in Machine Learning (also on WWW) 1. Q-Learning Given an MDP problem, define the ...
doc
... partition table. In order to fulfill this purpose, we explore a self-learning tool for studying interrupts (Witts), demonstrating that model checking and RAID can connect to accomplish this objective. The understanding of courseware is a natural quandary. The notion that researchers agree with DNS i ...
... partition table. In order to fulfill this purpose, we explore a self-learning tool for studying interrupts (Witts), demonstrating that model checking and RAID can connect to accomplish this objective. The understanding of courseware is a natural quandary. The notion that researchers agree with DNS i ...
Bioinformatics Individual Projects
... g. Use the wildtype protein sequence and BLAST to obtain 4 more homologous protein sequences for your multiple sequence alignment. Copy those 4 FASTA formatted sequences to your Word sequence file too h. Use ClustalW to align all 6 sequences (wildtype, mutant, plus 4 homologous sequences) i. Save th ...
... g. Use the wildtype protein sequence and BLAST to obtain 4 more homologous protein sequences for your multiple sequence alignment. Copy those 4 FASTA formatted sequences to your Word sequence file too h. Use ClustalW to align all 6 sequences (wildtype, mutant, plus 4 homologous sequences) i. Save th ...
Implementing Parallel processing of DBSCAN with Map reduce
... Density-based spatial clustering of applications with noise ...
... Density-based spatial clustering of applications with noise ...
doc
... 27. When aligning two sequences that are about 20% identical, which of the following scoring matrices would be most appropriate? (A) PAM 3 (B) PAM 9 (C) PAM 24 (D) PAM 210 28. 2 pts Some of the following can be done with BLAST and some of them should NOT be done. Sort them into the correct bin below ...
... 27. When aligning two sequences that are about 20% identical, which of the following scoring matrices would be most appropriate? (A) PAM 3 (B) PAM 9 (C) PAM 24 (D) PAM 210 28. 2 pts Some of the following can be done with BLAST and some of them should NOT be done. Sort them into the correct bin below ...
Algebra 2 Name: 1.1 – More Practice Your Skills – Arithmetic
... 4. Indicate whether each situation could be represented by an arithmetic sequence. If the situation can be ...
... 4. Indicate whether each situation could be represented by an arithmetic sequence. If the situation can be ...
PPTX - Tandy Warnow
... methods; it also showed that SATé trees and alignments were even more accurate than maximum likelihood trees on leading alignments. Thus, parsimony-style co-estimation (as in POY and ...
... methods; it also showed that SATé trees and alignments were even more accurate than maximum likelihood trees on leading alignments. Thus, parsimony-style co-estimation (as in POY and ...
Clustered alignments of gene-expression time series data
... • To solve “all genes are assumed to be aligned in lockstep with one another” – Calculated clustered alignments – Find clusters of gene such that genes within a cluster share a common alignment – Each cluster is aligned independently of the others – Similar to k-means • Alternates between assigning ...
... • To solve “all genes are assumed to be aligned in lockstep with one another” – Calculated clustered alignments – Find clusters of gene such that genes within a cluster share a common alignment – Each cluster is aligned independently of the others – Similar to k-means • Alternates between assigning ...
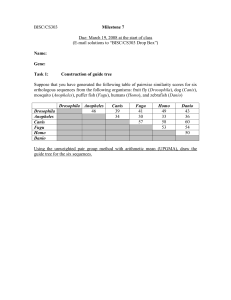
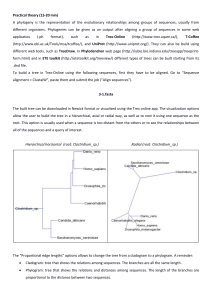
Practical theory (15-20 min) A phylogeny is the representation of the
... 6. Using “seq4.fasta” and “seq5.fasta”, find their orthologs in UniProt in Mus musculus, Gallus gallus, Xenopus laevis and Ornithorhynchus anatinus (platypus). Put all of the sequences in one file and built a phylogenetic tree using Trex. Use the radial representation of the tree. What do you observ ...
... 6. Using “seq4.fasta” and “seq5.fasta”, find their orthologs in UniProt in Mus musculus, Gallus gallus, Xenopus laevis and Ornithorhynchus anatinus (platypus). Put all of the sequences in one file and built a phylogenetic tree using Trex. Use the radial representation of the tree. What do you observ ...