Glossary - ChristopherKing.name
... RefSeq is a database of sequences that is edited by NCBI and is NON-redundant, meaning that it contains what NCBI determines is the strongest sequence data for each gene. Finally, we will be learning to use ClustalW, which is a multiple sequence alignment program. It allows you to enter a series of ...
... RefSeq is a database of sequences that is edited by NCBI and is NON-redundant, meaning that it contains what NCBI determines is the strongest sequence data for each gene. Finally, we will be learning to use ClustalW, which is a multiple sequence alignment program. It allows you to enter a series of ...
Introduction to BLAST ppt
... S. Needleman and C. Wunsch (1970). A general method applicable to the search for similarities in the amino acid sequence of two proteins, J. Molecular Biology, 48:443-453. D. Hirschberg (1975). A linear space algorithm for computing maximal common subsequences. Communications of the ACM, 18(6):341-3 ...
... S. Needleman and C. Wunsch (1970). A general method applicable to the search for similarities in the amino acid sequence of two proteins, J. Molecular Biology, 48:443-453. D. Hirschberg (1975). A linear space algorithm for computing maximal common subsequences. Communications of the ACM, 18(6):341-3 ...
LECTURE 1 INTRODUCTION Origin of word: Algorithm The word
... This model seems to go a good job of describing the computational power of most modern (nonparallel) machines. It does not model some elements, such as efficiency due to locality of reference, as described in the previous lecture. There are some “loop-holes” (or hid den ways of subverting the rules) ...
... This model seems to go a good job of describing the computational power of most modern (nonparallel) machines. It does not model some elements, such as efficiency due to locality of reference, as described in the previous lecture. There are some “loop-holes” (or hid den ways of subverting the rules) ...
biopatt - Carnegie Mellon School of Computer Science
... hidden Markov models. Pfam is available on the World Wide Web in the UK,…, Sweden, …, France, …, US. The latest version (6.6) of Pfam contains 3071 families, which match 69% of proteins in SWISS-PROT 39 and TrEMBL 14. Structural data, where available, have been utilised to ensure that Pfam families ...
... hidden Markov models. Pfam is available on the World Wide Web in the UK,…, Sweden, …, France, …, US. The latest version (6.6) of Pfam contains 3071 families, which match 69% of proteins in SWISS-PROT 39 and TrEMBL 14. Structural data, where available, have been utilised to ensure that Pfam families ...
Summary Team members: Weiqian Yan, Kanchan Khurad, and Yi
... The paper, A Monte Carlo Algorithm for Fast Projective Clustering, proposes 2 novel approaches to approximate optimal clusters in high dimensional data space. As research has proven, existing clustering methods that work well in low dimensional spaces don’t work well in high dimensional space due to ...
... The paper, A Monte Carlo Algorithm for Fast Projective Clustering, proposes 2 novel approaches to approximate optimal clusters in high dimensional data space. As research has proven, existing clustering methods that work well in low dimensional spaces don’t work well in high dimensional space due to ...
Supplementary Material (doc 28K)
... The complete set of parameters of TEIRESIAS used in this analysis is as follows: amino acids in the pattern (-l), number of overlapping characters in the convolved pattern (-c), maximum length of an elementary pattern (-w), minimum number of appearances of the pattern (-k), maximum number of bracket ...
... The complete set of parameters of TEIRESIAS used in this analysis is as follows: amino acids in the pattern (-l), number of overlapping characters in the convolved pattern (-c), maximum length of an elementary pattern (-w), minimum number of appearances of the pattern (-k), maximum number of bracket ...
File
... 1. Temperature readings are taken at 20 weather stations throughout the UK. Readings are taken at each station 8 times in one day. a) Describe how a 2-D array could be used to store the temperatures for each station. b) Declare this array c) Write an algorithm that will count the number of occasions ...
... 1. Temperature readings are taken at 20 weather stations throughout the UK. Readings are taken at each station 8 times in one day. a) Describe how a 2-D array could be used to store the temperatures for each station. b) Declare this array c) Write an algorithm that will count the number of occasions ...
OLD_s1a_alg_analysis..
... Worst-case running time of an algorithm: The longest running time for any input of size n An upper bound on the running time for any input guarantee that the algorithm will never take longer Example: Sort a set of numbers in increasing order; and the input is in decreasing order The wors ...
... Worst-case running time of an algorithm: The longest running time for any input of size n An upper bound on the running time for any input guarantee that the algorithm will never take longer Example: Sort a set of numbers in increasing order; and the input is in decreasing order The wors ...
What is Sequence Alignment?
... homology and those that occur by chance • Define a scoring function that accounts for mismatches and gaps Scoring Function (F): ...
... homology and those that occur by chance • Define a scoring function that accounts for mismatches and gaps Scoring Function (F): ...
Bioinformatics
... two fields have similar aims but the major difference is in scale. Bioinformatics deals with basic biological data and pays attention to details while Biological Computation is a subset of CS that builds large scale theoretical models of biological systems in an attempt to expand understanding of ...
... two fields have similar aims but the major difference is in scale. Bioinformatics deals with basic biological data and pays attention to details while Biological Computation is a subset of CS that builds large scale theoretical models of biological systems in an attempt to expand understanding of ...
Molecular Phylogenetic Analysis: Design and Implementation of
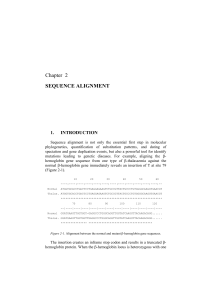
... selected, the first step is to align them [1,2]. The difference in lengths can appear due to sequencing errors (digitalizing the biological sample), mutations (insertions or deletions of one or more sites along the sequence) or because the researcher also wants to include fragments of the same genet ...
... selected, the first step is to align them [1,2]. The difference in lengths can appear due to sequencing errors (digitalizing the biological sample), mutations (insertions or deletions of one or more sites along the sequence) or because the researcher also wants to include fragments of the same genet ...