Exercise 11 - Understanding the Output for a blastn Search
... When we design a BLAST search, there are three basic decisions we must make: the BLAST program we wish to use, the query sequence we want to annotate, and the database we want to search. In addition, there are several optional parameters (such as the ‘expect’ threshold and other scoring parameters) ...
... When we design a BLAST search, there are three basic decisions we must make: the BLAST program we wish to use, the query sequence we want to annotate, and the database we want to search. In addition, there are several optional parameters (such as the ‘expect’ threshold and other scoring parameters) ...
Chapter 2: Fundamentals of the Analysis of Algorithm
... It cannot be investigated the way the previous examples are. ...
... It cannot be investigated the way the previous examples are. ...
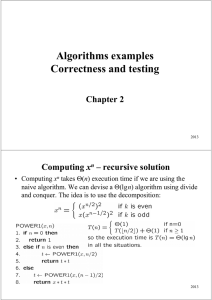
Algorithms examples Correctness and testing
... • This condition is called a loop invariant. • The proof is made by induction on the number i of executions of the body of the while loop. • If i=0 then the result is trivial because m = m0 and n = n0. • For the induction step we assume that at the end of the ith execution of the while loop we have ...
... • This condition is called a loop invariant. • The proof is made by induction on the number i of executions of the body of the while loop. • If i=0 then the result is trivial because m = m0 and n = n0. • For the induction step we assume that at the end of the ith execution of the while loop we have ...
Speeding up the Consensus Clustering methodology for microarray
... • NMF: Non negative Matrix Factorization, can also use another algorithm’s solution as a starting point(NMF, NMF-R, NMF-C, NMF-S) ...
... • NMF: Non negative Matrix Factorization, can also use another algorithm’s solution as a starting point(NMF, NMF-R, NMF-C, NMF-S) ...
Optimization of Aperiodically Spaced Antenna Arrays for Wideband
... simulates the “swarming” nature of bees when searching for food Semi-heuristic method Utilizes both local search and ...
... simulates the “swarming” nature of bees when searching for food Semi-heuristic method Utilizes both local search and ...
L10: k-Means Clustering
... • Random set of k points. By coupon collectors, we know that we need about k log k to get one in each cluster. • Randomly partition X = {X1 , X2 , . . . , Xk } and take ci = average(Xi ). This biases towards “center” of X (by Chernoff-Hoeffding). • Gonzalez algorithm (for k-center). This biases too ...
... • Random set of k points. By coupon collectors, we know that we need about k log k to get one in each cluster. • Randomly partition X = {X1 , X2 , . . . , Xk } and take ci = average(Xi ). This biases towards “center” of X (by Chernoff-Hoeffding). • Gonzalez algorithm (for k-center). This biases too ...
Full text
... function (1 - x - x2 - ••• - xk)~l was found by V. Schlegel in 1894. See [1, Chap. XVII] for this and other classical references. An alternate solution to the problem can be obtained as follows. Consider a sequence of experiments: Toss a p-coin Xl times, until a sequence of k - 1 heads occurs. Then ...
... function (1 - x - x2 - ••• - xk)~l was found by V. Schlegel in 1894. See [1, Chap. XVII] for this and other classical references. An alternate solution to the problem can be obtained as follows. Consider a sequence of experiments: Toss a p-coin Xl times, until a sequence of k - 1 heads occurs. Then ...
PPTX - UT Computer Science
... Metagenomic Taxon Identification Objective: classify short reads in a metagenomic sample ...
... Metagenomic Taxon Identification Objective: classify short reads in a metagenomic sample ...
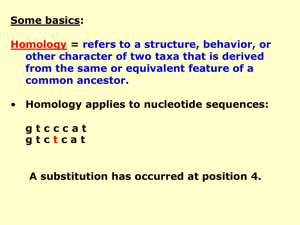
String-Matching Problem
... Rabin-Karp Algorithm Correctness: T is a string of
characters over an alphabet of size d, P
is string of characters over an alphabet of
size d and |P| <= |T|, d is the size of the
alphabet and q is a prime number
...
... Rabin-Karp Algorithm Correctness
Slides 4 - UF CISE - University of Florida
... • Different levels of the BLOSUM matrix can be created by differentially weighting the degree of similarity between sequences. For example, a BLOSUM62 matrix is calculated from protein blocks such that if two sequences are more than 62% identical, then the contribution of these sequences is weighted ...
... • Different levels of the BLOSUM matrix can be created by differentially weighting the degree of similarity between sequences. For example, a BLOSUM62 matrix is calculated from protein blocks such that if two sequences are more than 62% identical, then the contribution of these sequences is weighted ...
Greedy Algorithms
... – If there is no decreasing strip, there may be no reversal r that reduces the number of breakpoints (i.e. b(p• r) ≥ b(p) for any reversal r). – By reversing an increasing strip ( # of breakpoints stay unchanged ), we will create a decreasing strip at the next step. Then the number of breakpoints w ...
... – If there is no decreasing strip, there may be no reversal r that reduces the number of breakpoints (i.e. b(p• r) ≥ b(p) for any reversal r). – By reversing an increasing strip ( # of breakpoints stay unchanged ), we will create a decreasing strip at the next step. Then the number of breakpoints w ...
Bioinformatics with basic local alignment search tool (BLAST) and
... 1988). There are three main types of sequence alignments: pairwise sequence alignment, multiple sequence alignment and structural sequence alignment (Pearson and Lipman, 1988; Luscombe et al., 2001). Pairwise sequence alignment can only be used between two sequences at a time. Multiple sequence alig ...
... 1988). There are three main types of sequence alignments: pairwise sequence alignment, multiple sequence alignment and structural sequence alignment (Pearson and Lipman, 1988; Luscombe et al., 2001). Pairwise sequence alignment can only be used between two sequences at a time. Multiple sequence alig ...
Greedy Algorithms - University of Illinois at Urbana
... • ImprovedBreakPointReversalSort is an approximation algorithm with a performance guarantee of at most 4 – It eliminates at least one breakpoint in every two steps; at most 2b(p) steps – Approximation ratio: 2b(p) / d(p) – Optimal algorithm eliminates at most 2 breakpoints in every step: d(p) b(p) ...
... • ImprovedBreakPointReversalSort is an approximation algorithm with a performance guarantee of at most 4 – It eliminates at least one breakpoint in every two steps; at most 2b(p) steps – Approximation ratio: 2b(p) / d(p) – Optimal algorithm eliminates at most 2 breakpoints in every step: d(p) b(p) ...