Lecture 1
... Calculations show that proteins are NOT very stable molecules Net stabilization of ~0.4 kJ mol-1 for each amino acid ...
... Calculations show that proteins are NOT very stable molecules Net stabilization of ~0.4 kJ mol-1 for each amino acid ...
Supplementary Table 1: A complete list of proteins identified with
... culture) analysis of MOLM-13 cells treated with nutlin-3 (6 µM, 6h) is provided. MOLM-13 cells treated with DMSO (control) were labeled with light isotopes of amino acids (L), and nutlin-treated cells were labeled with heavy isotopes of amino acids (H); the reported regulation of proteins in respons ...
... culture) analysis of MOLM-13 cells treated with nutlin-3 (6 µM, 6h) is provided. MOLM-13 cells treated with DMSO (control) were labeled with light isotopes of amino acids (L), and nutlin-treated cells were labeled with heavy isotopes of amino acids (H); the reported regulation of proteins in respons ...
Capturing denaturing proteins * Small Heat Shock Protein substrate
... sHSP chaperone action and interaction with substrates, therefore, has wide-ranging implications for understanding cellular stress and disease processes. We are studying the mechanism of sHSP substrate recognition by identifying specific crosslinking sites between sHSPs and denaturing substrates. sHS ...
... sHSP chaperone action and interaction with substrates, therefore, has wide-ranging implications for understanding cellular stress and disease processes. We are studying the mechanism of sHSP substrate recognition by identifying specific crosslinking sites between sHSPs and denaturing substrates. sHS ...
Knuffke Prezi- Macromolecules
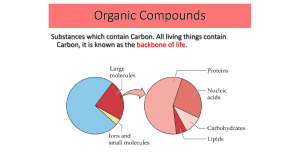
... Organic Compounds Substances which contain Carbon. All living things contain Carbon, it is known as the backbone of life. ...
... Organic Compounds Substances which contain Carbon. All living things contain Carbon, it is known as the backbone of life. ...
Essential amino acids and nutrition
... 1. Consider why protein is needed and what amino acids and proteins are used for in the body. Describe the symptoms you would expect a person with protein deficiency to have. ...
... 1. Consider why protein is needed and what amino acids and proteins are used for in the body. Describe the symptoms you would expect a person with protein deficiency to have. ...
File - SMIC Nutrition Science
... 6. The making of protein from 20 amino acids was compared in the chapter to the use of the English alphabet (26 letters) to make words and speak the English language. Why was this such a fitting ...
... 6. The making of protein from 20 amino acids was compared in the chapter to the use of the English alphabet (26 letters) to make words and speak the English language. Why was this such a fitting ...
Two Rules on Protein-Ligand Interactions Xiaodong Pang1, 2
... Understanding the ruling principles of interaction between a target protein and a ligand is of paramount importance in drug discovery efforts. So far, in finding a real ligand for a given target protein, we are limited to experimental screening from a large number of small molecules, or through free ...
... Understanding the ruling principles of interaction between a target protein and a ligand is of paramount importance in drug discovery efforts. So far, in finding a real ligand for a given target protein, we are limited to experimental screening from a large number of small molecules, or through free ...
Proteins - Wesleyan College Faculty
... http://learn.genetics.utah.edu/content/begin/dna/transcribe/ ...
... http://learn.genetics.utah.edu/content/begin/dna/transcribe/ ...
November 19, 2012 3:00 PM Livermore Center 101 Isaac C. Sanchez
... Within a polymer thin film, free-volume elements have a wide range of size and topology. This broad range of free-volume element sizes determines the ability for a polymer to perform molecular separations. Using atomistic models, cavity size (free volume) distributions were determined by a combinati ...
... Within a polymer thin film, free-volume elements have a wide range of size and topology. This broad range of free-volume element sizes determines the ability for a polymer to perform molecular separations. Using atomistic models, cavity size (free volume) distributions were determined by a combinati ...
Protein: How Cows and Carrots Become People 1. Your body can
... 9. Believe it or not, you have hydrochloric acid in your stomach. It’s nasty stuff that can burn skin and eat away at almost anything. What is it doing in your stomach and why ...
... 9. Believe it or not, you have hydrochloric acid in your stomach. It’s nasty stuff that can burn skin and eat away at almost anything. What is it doing in your stomach and why ...
Faraday Discussion Meeting September 2002
... The effect of applied force on the energy landscape that describes protein conformation is an exciting and challenging topic in molecular biophysics. Recently it has become possible to use nanotechnology tools such as the atomic force microscope and laser tweezers to manipulate individual molecules ...
... The effect of applied force on the energy landscape that describes protein conformation is an exciting and challenging topic in molecular biophysics. Recently it has become possible to use nanotechnology tools such as the atomic force microscope and laser tweezers to manipulate individual molecules ...
4.9.teaching.notes
... Learning Objectives Explain the process of protein folding using appropriate terminology. Describe the structure of insulin. ...
... Learning Objectives Explain the process of protein folding using appropriate terminology. Describe the structure of insulin. ...
DOC
... deGradFP harnesses the ubiquitin-proteasome pathway to achieve direct depletion of GFP-tagged proteins. deGradFP is in essence a universal method because it relies on an evolutionarily conserved machinery for protein catabolism in eukaryotic cells; see refs. 5, 6 for review. deGradFP is particularly ...
... deGradFP harnesses the ubiquitin-proteasome pathway to achieve direct depletion of GFP-tagged proteins. deGradFP is in essence a universal method because it relies on an evolutionarily conserved machinery for protein catabolism in eukaryotic cells; see refs. 5, 6 for review. deGradFP is particularly ...
Visually Demonstrating the Principles of Protein Folding
... Mathematical model uses amino acid sequences and these values to predict secondary structure. ...
... Mathematical model uses amino acid sequences and these values to predict secondary structure. ...
PROTEIN STRUCTURE ANALYSIS ASSIGNMENTS Search from
... How many structures do you find? What methods have been used for their determination? How do they differ? Which one do you choose and why? Is it a crystallographic or NMR structure? What is resolution? How many chains are on the structure?What is the number of amino acids in the structure? What is t ...
... How many structures do you find? What methods have been used for their determination? How do they differ? Which one do you choose and why? Is it a crystallographic or NMR structure? What is resolution? How many chains are on the structure?What is the number of amino acids in the structure? What is t ...
Center for Structural Biology
... Review: Heirarchy of Structure Primary- sequence Secondary- local Supersecondary (motifs)- intermediate Domains- independent folding units Tertiary- organization of a complete chain Quaternary- organization of multiple chains ...
... Review: Heirarchy of Structure Primary- sequence Secondary- local Supersecondary (motifs)- intermediate Domains- independent folding units Tertiary- organization of a complete chain Quaternary- organization of multiple chains ...
hw1009-aminoacids-proteins
... In this video, we see molecules hooking together to form macromolecules. The molecule is an amino acid or peptide, joining together to form a poplypeptide or protein. Please answer these questions: ...
... In this video, we see molecules hooking together to form macromolecules. The molecule is an amino acid or peptide, joining together to form a poplypeptide or protein. Please answer these questions: ...
Quiz-2
... 5. The helical structure in collagen is very compact. What is the structural explanation for this phenomenon? 6. What were the two physical criterion used by Ramchandran to predict allowed conformations? 7. Ramchandran’s plot for poly-glycine shows four big squares of allowed conformations which is ...
... 5. The helical structure in collagen is very compact. What is the structural explanation for this phenomenon? 6. What were the two physical criterion used by Ramchandran to predict allowed conformations? 7. Ramchandran’s plot for poly-glycine shows four big squares of allowed conformations which is ...
Proceedings of a meeting held at Allerton House, Monticello, Illinois
... these angles to better than a tenth of a radian, there would be 10300 possible configurations in our theoretical protein. In nature, proteins apparently do not sample all of these possible configurations since they fold in a few seconds, and even postulating a minimum time for going from one conform ...
... these angles to better than a tenth of a radian, there would be 10300 possible configurations in our theoretical protein. In nature, proteins apparently do not sample all of these possible configurations since they fold in a few seconds, and even postulating a minimum time for going from one conform ...
Combinatorial docking approach for structure prediction of large
... for pair-wise docking of smaller molecules or protein domains as well as free-energy calculations to build determine native conformations of larger protein complexes. Such an algorithm is much faster than trying to determine the entire protein or assembly structure through molecular dynamics methods ...
... for pair-wise docking of smaller molecules or protein domains as well as free-energy calculations to build determine native conformations of larger protein complexes. Such an algorithm is much faster than trying to determine the entire protein or assembly structure through molecular dynamics methods ...
Estimation of the protein secondary structure in aqueous solutions
... The secondary structure of proteins is very important for their proper functioning. The investigation of the secondary structure gives us an insight into the mechanisms of protein functioning in the living cell. IR absorption spectroscopy provides the opportunity to identify a large number of types ...
... The secondary structure of proteins is very important for their proper functioning. The investigation of the secondary structure gives us an insight into the mechanisms of protein functioning in the living cell. IR absorption spectroscopy provides the opportunity to identify a large number of types ...
Reading Guide: Pratt and Cornely, Chapter 4, pp 87
... 14. Draw a parallel beta sheet between two oligonucleotides that are five alanine residues long. How is an antiparallel sheet different in h-bonding? 15. What is an irregular secondary structure? What is a loop? 16. Define a protein domain. 17. Why is the interior of a globular protein often regular ...
... 14. Draw a parallel beta sheet between two oligonucleotides that are five alanine residues long. How is an antiparallel sheet different in h-bonding? 15. What is an irregular secondary structure? What is a loop? 16. Define a protein domain. 17. Why is the interior of a globular protein often regular ...
Protein Study Guide
... 3. Sketch two amino acids side-by-side, on one of them label the alpha or central carbon, amino, carboxyl, then show how the two can be joined together. ...
... 3. Sketch two amino acids side-by-side, on one of them label the alpha or central carbon, amino, carboxyl, then show how the two can be joined together. ...
Rosetta@home

Rosetta@home is a distributed computing project for protein structure prediction on the Berkeley Open Infrastructure for Network Computing (BOINC) platform, run by the Baker laboratory at the University of Washington. Rosetta@home aims to predict protein–protein docking and design new proteins with the help of about sixty thousand active volunteered computers processing at 83 teraFLOPS on average as of April 18, 2014. Foldit, a Rosetta@Home videogame, aims to reach these goals with a crowdsourcing approach. Though much of the project is oriented towards basic research on improving the accuracy and robustness of the proteomics methods, Rosetta@home also does applied research on malaria, Alzheimer's disease and other pathologies.Like all BOINC projects, Rosetta@home uses idle computer processing resources from volunteers' computers to perform calculations on individual workunits. Completed results are sent to a central project server where they are validated and assimilated into project databases. The project is cross-platform, and runs on a wide variety of hardware configurations. Users can view the progress of their individual protein structure prediction on the Rosetta@home screensaver.In addition to disease-related research, the Rosetta@home network serves as a testing framework for new methods in structural bioinformatics. These new methods are then used in other Rosetta-based applications, like RosettaDock and the Human Proteome Folding Project, after being sufficiently developed and proven stable on Rosetta@home's large and diverse collection of volunteer computers. Two particularly important tests for the new methods developed in Rosetta@home are the Critical Assessment of Techniques for Protein Structure Prediction (CASP) and Critical Assessment of Prediction of Interactions (CAPRI) experiments, biannual experiments which evaluate the state of the art in protein structure prediction and protein–protein docking prediction, respectively. Rosetta@home consistently ranks among the foremost docking predictors, and is one of the best tertiary structure predictors available.