Proteins
... 6. Label the type of bond used to make proteins. 7. Draw arrows to identify these bonds in your model 8. Label the N-terminus and C-terminus 9. Put SQUARES around the R groups 10. Use your amino acid chart to identify & label the type of R group (non-polar, polar, charge basic, charged acidic, etc) ...
... 6. Label the type of bond used to make proteins. 7. Draw arrows to identify these bonds in your model 8. Label the N-terminus and C-terminus 9. Put SQUARES around the R groups 10. Use your amino acid chart to identify & label the type of R group (non-polar, polar, charge basic, charged acidic, etc) ...
more details
... Abstract: Standard models of protein evolution generally assume each location evolves independently, although it is well appreciated that substitution rates in a protein are influenced by changes in the amino acids at other locations. Generating more accurate but computationally tractable models of ...
... Abstract: Standard models of protein evolution generally assume each location evolves independently, although it is well appreciated that substitution rates in a protein are influenced by changes in the amino acids at other locations. Generating more accurate but computationally tractable models of ...
The measurement of the biological inventory of proteins within an
... metaproteomics for its inclusion of a biological community, it has shown potential to significantly improve the understanding of ocean ecology and biogeochemistry by allowing a broad diagnosis of ecosystems across large geographical distances. In this manner the measurement of proteins has great po ...
... metaproteomics for its inclusion of a biological community, it has shown potential to significantly improve the understanding of ocean ecology and biogeochemistry by allowing a broad diagnosis of ecosystems across large geographical distances. In this manner the measurement of proteins has great po ...
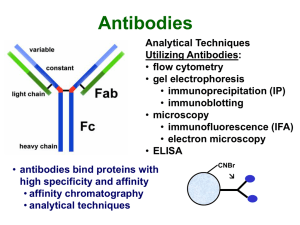
Typical IP Protocol
... • also provides information about synthesis, posttranslational events, etc. ...
... • also provides information about synthesis, posttranslational events, etc. ...
Presentazione standard di PowerPoint
... flexibility and temporal evolution in the analysis of molecular systems. During these years I have been involved in the study of many different proteins with potential application in biotechnology. The knowledge of their dynamics using a MD approach, coupled to the experimental evaluation of their s ...
... flexibility and temporal evolution in the analysis of molecular systems. During these years I have been involved in the study of many different proteins with potential application in biotechnology. The knowledge of their dynamics using a MD approach, coupled to the experimental evaluation of their s ...
Protein Structure and Folding
... contains 8 cysteine residues that form 4 four bridges. Derive expression for the number of ways N cysteins can pair with each other. Some proteins like many neutrophic factors contain odd number of cysteins. What kind of pair would you expect in this case? 2. Rearrange equation H - TS = RTln[D]/[N ...
... contains 8 cysteine residues that form 4 four bridges. Derive expression for the number of ways N cysteins can pair with each other. Some proteins like many neutrophic factors contain odd number of cysteins. What kind of pair would you expect in this case? 2. Rearrange equation H - TS = RTln[D]/[N ...
Protein Structure and Folding
... 1. Use SCOP (Structural Classification Of Proteins) http://scop.mrc-lmb.cam.ac.uk/scop/ to classify PDB entry 1tml. 2. Name the fold of central domain of 1m6h and draw the corresponding topology diagram. 3. Classify the two domains of a metabolic regulator protein 1d66 from Baker’s yeast. 4. Use DAL ...
... 1. Use SCOP (Structural Classification Of Proteins) http://scop.mrc-lmb.cam.ac.uk/scop/ to classify PDB entry 1tml. 2. Name the fold of central domain of 1m6h and draw the corresponding topology diagram. 3. Classify the two domains of a metabolic regulator protein 1d66 from Baker’s yeast. 4. Use DAL ...
Proteins for Growth and Repair
... Too much protein can harm your body. If you eat too much protein the excess will be stored as body fat. Over-consumption of protein can also cause constipation, diarrhea, excessive gas, dehydration, dizziness and bad breath. ...
... Too much protein can harm your body. If you eat too much protein the excess will be stored as body fat. Over-consumption of protein can also cause constipation, diarrhea, excessive gas, dehydration, dizziness and bad breath. ...
PLANT PROTEINS FOR THE FUTURE-English
... pigeon pea, etc. are currently the most important legumes for human consumption and animal feed. Amaranth and quinoa are considered “pseudocereals” and are also good sources of proteins. Amaranth seeds contain lysine, an essential amino acid, limited in other grains or plant sources but are limited ...
... pigeon pea, etc. are currently the most important legumes for human consumption and animal feed. Amaranth and quinoa are considered “pseudocereals” and are also good sources of proteins. Amaranth seeds contain lysine, an essential amino acid, limited in other grains or plant sources but are limited ...
Yellow Neuphoria - Controlled Labs
... craves tasty protein. In the past that has been the dilemma. PROnom 23™ solves the fight between muscle and stomach by offering both the highest quality protein and an amazing dessert like taste. Once you try PROnom 23™ for the first time you will never go back to your old protein powder again; it’s ...
... craves tasty protein. In the past that has been the dilemma. PROnom 23™ solves the fight between muscle and stomach by offering both the highest quality protein and an amazing dessert like taste. Once you try PROnom 23™ for the first time you will never go back to your old protein powder again; it’s ...
ppt
... • There are 20 amino acids • Each amino acid has a central carbon atom, Ca • Each Ca has an attached amino group and a carboxyl group • Each Ca has an attached side chain • Adjacent peptides link through a ...
... • There are 20 amino acids • Each amino acid has a central carbon atom, Ca • Each Ca has an attached amino group and a carboxyl group • Each Ca has an attached side chain • Adjacent peptides link through a ...
Slide 1
... The kinetic Theory of Protein Folding Folding proceeds through a definite series of steps or a Pathway. A protein does not try out all possible rotations of conformational angles, but only enough to find the pathway. ...
... The kinetic Theory of Protein Folding Folding proceeds through a definite series of steps or a Pathway. A protein does not try out all possible rotations of conformational angles, but only enough to find the pathway. ...
The World of Chemistry Episode 24
... There are four subunits, each containing 2 - helices and 2 - sheets. An atom of iron is found in the center of each. 3. Briefly describe the four types of protein structure. Primary - the sequence of amino acids in the protein chain Secondary - the formation of - helices or - sheets from the ...
... There are four subunits, each containing 2 - helices and 2 - sheets. An atom of iron is found in the center of each. 3. Briefly describe the four types of protein structure. Primary - the sequence of amino acids in the protein chain Secondary - the formation of - helices or - sheets from the ...
Episode 24 - The Genetic Code
... There are four subunits, each containing 2 - helices and 2 - sheets. An atom of iron is found in the center of each. 3. Briefly describe the four types of protein structure. Primary - the sequence of amino acids in the protein chain Secondary - the formation of - helices or - sheets from the ...
... There are four subunits, each containing 2 - helices and 2 - sheets. An atom of iron is found in the center of each. 3. Briefly describe the four types of protein structure. Primary - the sequence of amino acids in the protein chain Secondary - the formation of - helices or - sheets from the ...
Survey of Protein Structure Prediction Methods
... Trying to predict the end result of folding, using a large amount of comparison between known and unknown structures Protein folding problem Trying to understand the folding path which leads to the end result of folding, typically by MD simulations or energy calculation ...
... Trying to predict the end result of folding, using a large amount of comparison between known and unknown structures Protein folding problem Trying to understand the folding path which leads to the end result of folding, typically by MD simulations or energy calculation ...
G Protein Coupled Receptors
... Schematic drawing of the flow of events in G protein activation. The process starts when the Arg of the DRY motif moves toward the cytosol. The signal moves `through' helix 5 to the nucleotide binding site. The GDP– GTP exchange leads to a conformational change in the switch region. This leads to di ...
... Schematic drawing of the flow of events in G protein activation. The process starts when the Arg of the DRY motif moves toward the cytosol. The signal moves `through' helix 5 to the nucleotide binding site. The GDP– GTP exchange leads to a conformational change in the switch region. This leads to di ...
slides
... These methods were based on the helix- or sheet-forming propensities of individual amino acids, sometimes coupled with rules for estimating the free energy of forming secondary structure elements. Such methods were typically ~60% accurate in predicting which of the three states (helix/sheet/coil) a ...
... These methods were based on the helix- or sheet-forming propensities of individual amino acids, sometimes coupled with rules for estimating the free energy of forming secondary structure elements. Such methods were typically ~60% accurate in predicting which of the three states (helix/sheet/coil) a ...
Questions for Discussion or Assignment to Accompany the Ubiquitin
... This assumes that the amplifier is linear and assumes that PL2 and PL1 represent attenuations (i.e. higher attenuation is less power). (c) Now calculate the 360 degree pulse at PL2. (d) Assuming a presaturation pulse of 4 seconds, how many revolutions does the water magnetization vector undergo if ...
... This assumes that the amplifier is linear and assumes that PL2 and PL1 represent attenuations (i.e. higher attenuation is less power). (c) Now calculate the 360 degree pulse at PL2. (d) Assuming a presaturation pulse of 4 seconds, how many revolutions does the water magnetization vector undergo if ...
View attached file
... In various cases such misfolding can be reversible under experimental conditions. Thus, drugs that will cause refolding of the misfolded protein may restore its function and prevent its harmful aggregation. Research methods: In vitro and bacterial assays of protein folding and aggregation (in collab ...
... In various cases such misfolding can be reversible under experimental conditions. Thus, drugs that will cause refolding of the misfolded protein may restore its function and prevent its harmful aggregation. Research methods: In vitro and bacterial assays of protein folding and aggregation (in collab ...
D6- Bulletin Board Powerful Protein
... • There are 9 essential amino acids that our bodies can’t make, so we need to get them from our food. • If a protein food has all 9 essential amino acids, it is called a complete protein. If it doesn’t, it is called an incomplete protein. • You can eat incomplete protein foods together to make sure ...
... • There are 9 essential amino acids that our bodies can’t make, so we need to get them from our food. • If a protein food has all 9 essential amino acids, it is called a complete protein. If it doesn’t, it is called an incomplete protein. • You can eat incomplete protein foods together to make sure ...
Rosetta@home

Rosetta@home is a distributed computing project for protein structure prediction on the Berkeley Open Infrastructure for Network Computing (BOINC) platform, run by the Baker laboratory at the University of Washington. Rosetta@home aims to predict protein–protein docking and design new proteins with the help of about sixty thousand active volunteered computers processing at 83 teraFLOPS on average as of April 18, 2014. Foldit, a Rosetta@Home videogame, aims to reach these goals with a crowdsourcing approach. Though much of the project is oriented towards basic research on improving the accuracy and robustness of the proteomics methods, Rosetta@home also does applied research on malaria, Alzheimer's disease and other pathologies.Like all BOINC projects, Rosetta@home uses idle computer processing resources from volunteers' computers to perform calculations on individual workunits. Completed results are sent to a central project server where they are validated and assimilated into project databases. The project is cross-platform, and runs on a wide variety of hardware configurations. Users can view the progress of their individual protein structure prediction on the Rosetta@home screensaver.In addition to disease-related research, the Rosetta@home network serves as a testing framework for new methods in structural bioinformatics. These new methods are then used in other Rosetta-based applications, like RosettaDock and the Human Proteome Folding Project, after being sufficiently developed and proven stable on Rosetta@home's large and diverse collection of volunteer computers. Two particularly important tests for the new methods developed in Rosetta@home are the Critical Assessment of Techniques for Protein Structure Prediction (CASP) and Critical Assessment of Prediction of Interactions (CAPRI) experiments, biannual experiments which evaluate the state of the art in protein structure prediction and protein–protein docking prediction, respectively. Rosetta@home consistently ranks among the foremost docking predictors, and is one of the best tertiary structure predictors available.