Using a Mechanistic Perspective to Simulate Protein Backbone Motion
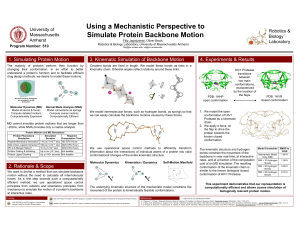
... conformation of the kinematic chain is similar to the known biological closed conformation of HIV1 Protease. ...
... conformation of the kinematic chain is similar to the known biological closed conformation of HIV1 Protease. ...
Document
... 1) Forms protein’s initial S-S bonds in similar way (protein –SH attacks PDI S-S bond to give mixed disulfide) 2) Protein SH attacks protein-PDI mixed S-S bond to give protein S-S bond 3) Continues until protein in native S-S configuration and PDI cannot bind to exposed hydrophobic patches on the pr ...
... 1) Forms protein’s initial S-S bonds in similar way (protein –SH attacks PDI S-S bond to give mixed disulfide) 2) Protein SH attacks protein-PDI mixed S-S bond to give protein S-S bond 3) Continues until protein in native S-S configuration and PDI cannot bind to exposed hydrophobic patches on the pr ...
Protein Folding
... • Protein folding considers the question of how the process of protein folding occurs, i. e. unfolded native state. • This very challenging problem has been described as the second half of the genetic code, and as the three-dimensional code, as opposed to the one-dimensional code involved in nucle ...
... • Protein folding considers the question of how the process of protein folding occurs, i. e. unfolded native state. • This very challenging problem has been described as the second half of the genetic code, and as the three-dimensional code, as opposed to the one-dimensional code involved in nucle ...
Protein Domains
... When the amino acid sequences of two proteins are compared and found to exhibit significant similarity they are assumed to be evolutionarily related i.e. they are homologues two classes of homologue (orthologue and paralogue) orthologous genes are descended from a unique ancestral gene and their di ...
... When the amino acid sequences of two proteins are compared and found to exhibit significant similarity they are assumed to be evolutionarily related i.e. they are homologues two classes of homologue (orthologue and paralogue) orthologous genes are descended from a unique ancestral gene and their di ...
John Torri Basic Nutrition Special Topic: Protein November 13 2014
... As we have learned from our Nutrition class, we need a daily intake of carbohydrates, fats, and proteins. Most people don’t know how proteins are stored, sources of proteins, or even how they work. I found an article that helps shed light on this topic. According to “Choosing Protein Wisely” Our bod ...
... As we have learned from our Nutrition class, we need a daily intake of carbohydrates, fats, and proteins. Most people don’t know how proteins are stored, sources of proteins, or even how they work. I found an article that helps shed light on this topic. According to “Choosing Protein Wisely” Our bod ...
What is PCM Synergy? PCM synergy is a quality blend is a multi
... What is PCM Synergy? PCM synergy is a quality blend is a multi-functional protein supplement that guarantees a good supply of protein building blocks for the individual. Furthermore, PCM synergy contains a high level of BCAAs (branch chain amino acids) which are vital in the manufacture, maintenance ...
... What is PCM Synergy? PCM synergy is a quality blend is a multi-functional protein supplement that guarantees a good supply of protein building blocks for the individual. Furthermore, PCM synergy contains a high level of BCAAs (branch chain amino acids) which are vital in the manufacture, maintenance ...
Document
... • Threading: Align sequence to structure (templates) For each alignment, the probability that that each amino acid residue would occur in such an environment is calculated based on observed preferences in determined structures. § Rationale: • Limited number of basic folds found in nature • Amino aci ...
... • Threading: Align sequence to structure (templates) For each alignment, the probability that that each amino acid residue would occur in such an environment is calculated based on observed preferences in determined structures. § Rationale: • Limited number of basic folds found in nature • Amino aci ...
6th semester-2006 Project Proposal
... “Construction of plasmid vectors to tag proteins for universal light-induced protein immobilization on surfaces” Background: A method of light-induced immobilization of proteins(1,2) on chemically treated surfaces has been successfully developed over the past years in the group, by Teresa Petersen a ...
... “Construction of plasmid vectors to tag proteins for universal light-induced protein immobilization on surfaces” Background: A method of light-induced immobilization of proteins(1,2) on chemically treated surfaces has been successfully developed over the past years in the group, by Teresa Petersen a ...
EGEE07_FP_October1st2007
... Protein function is linked to the specific three-dimensional arrangement of amino acids functional groups. With the advancement of molecular biology techniques a huge amount of information on protein sequences has been made available but far less information is available on structure and functio ...
... Protein function is linked to the specific three-dimensional arrangement of amino acids functional groups. With the advancement of molecular biology techniques a huge amount of information on protein sequences has been made available but far less information is available on structure and functio ...
Protein Kinases
... The reversible addition of phosphate groups to proteins is important for the transmission of signals within eukaryotic cells and, as a result, protein phosphorylation and dephosphorylation regulate many diverse cellular processes. As the number of known protein kinases has increased at an ever-accel ...
... The reversible addition of phosphate groups to proteins is important for the transmission of signals within eukaryotic cells and, as a result, protein phosphorylation and dephosphorylation regulate many diverse cellular processes. As the number of known protein kinases has increased at an ever-accel ...
An Approach to Including Protein Quality When
... The production of protein from animal sources is often criticized because of the low efficiency of converting plant protein from feeds into protein in the animal products. However, this critique does not consider the fact that large portions of the plant-based proteins fed to animals may be human-in ...
... The production of protein from animal sources is often criticized because of the low efficiency of converting plant protein from feeds into protein in the animal products. However, this critique does not consider the fact that large portions of the plant-based proteins fed to animals may be human-in ...
Discussion Problem Set 3 C483 Spring 2014
... Discussion Problem Set 3 C483 Spring 2014 Problems from lectures 6-8. (Through Jan 31) 1. What are the two main forces that stabilize an alpha helix? Describe them. How might you test the importance of an n-pi-star interaction? 2. What two things need to be minimized to have a stable alpha helix? 3. ...
... Discussion Problem Set 3 C483 Spring 2014 Problems from lectures 6-8. (Through Jan 31) 1. What are the two main forces that stabilize an alpha helix? Describe them. How might you test the importance of an n-pi-star interaction? 2. What two things need to be minimized to have a stable alpha helix? 3. ...
Slide 1
... – Retinal ligand shows binding pocket – In some 7TM proteins: different binding site ...
... – Retinal ligand shows binding pocket – In some 7TM proteins: different binding site ...
Metal Regulation and Signalling - Zn Proteins
... Structural Zn proteins have different functions, but all create interfaces for macromolecular interactions: protein-protein, protein-DNA, protein-RNA, protein-polysaccharides. No Zn binding motifs found in the e. coli genome, in contrast to an abundance of motifs in eukaryotic cells. Speculation tha ...
... Structural Zn proteins have different functions, but all create interfaces for macromolecular interactions: protein-protein, protein-DNA, protein-RNA, protein-polysaccharides. No Zn binding motifs found in the e. coli genome, in contrast to an abundance of motifs in eukaryotic cells. Speculation tha ...
CHEM F654
... acceptable, documented reason such as unexpected illness, family emergencies or other unavoidable events. The final exam could be an exclusive oral exam if students unanimously opt for this exam type. Presentations: Students will receive adequate preparation time for all assignments. Content and org ...
... acceptable, documented reason such as unexpected illness, family emergencies or other unavoidable events. The final exam could be an exclusive oral exam if students unanimously opt for this exam type. Presentations: Students will receive adequate preparation time for all assignments. Content and org ...
Purified Sp1 protein
... gene expression in the early development of an organism. The protein is 785 amino acids long, with a molecular weight of 81 kDa. The SP1 transcription factor contains a zinc finger protein motif, by which it binds directly to DNA and enhances gene transcription. Its zinc fingers are of the Cys2/His2 ...
... gene expression in the early development of an organism. The protein is 785 amino acids long, with a molecular weight of 81 kDa. The SP1 transcription factor contains a zinc finger protein motif, by which it binds directly to DNA and enhances gene transcription. Its zinc fingers are of the Cys2/His2 ...
Lecture 13_summary
... which can be related to amyloidosis (5-10 different sequence/predicted structural features) For example Fact : Amyloids tend to aggregare via beta sheet – Calculate: the percent of secondary structure (H,E, C) Fact : Amyloids tend to aggregate via aromatic residues Calculate : the percent of differe ...
... which can be related to amyloidosis (5-10 different sequence/predicted structural features) For example Fact : Amyloids tend to aggregare via beta sheet – Calculate: the percent of secondary structure (H,E, C) Fact : Amyloids tend to aggregate via aromatic residues Calculate : the percent of differe ...
Paper background for Students
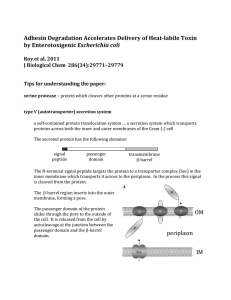
... The resulting fusion protein contains three domains: a. EtpA b. 10 amino acids of the myc protein sequence (a protein “tag) c. 6 histidine residues (a protein “tag”) This is useful because the protein can be purified using immobilized antibodies against the myc tag or the polyhistidine tag. Alternat ...
... The resulting fusion protein contains three domains: a. EtpA b. 10 amino acids of the myc protein sequence (a protein “tag) c. 6 histidine residues (a protein “tag”) This is useful because the protein can be purified using immobilized antibodies against the myc tag or the polyhistidine tag. Alternat ...
... “We knew the sequenator had a much greater potential so we delayed publication of the complete paper for another three years until the performance was improved. “The paper is highly cited because, until the invention of the sequenator, no single technique for sequencing was universally accepted. The ...
The molecular architecture, macro-organization and functions of the
... stabilization of the ultrastructure of thylakoid membranes and in their reorganizations. In addition, these proteins play key roles in important regulatory mechanisms: in excess light, via controlled dissipation governed by low lumenal pH, they are capable of transiently downregulating their light-h ...
... stabilization of the ultrastructure of thylakoid membranes and in their reorganizations. In addition, these proteins play key roles in important regulatory mechanisms: in excess light, via controlled dissipation governed by low lumenal pH, they are capable of transiently downregulating their light-h ...
Rosetta@home

Rosetta@home is a distributed computing project for protein structure prediction on the Berkeley Open Infrastructure for Network Computing (BOINC) platform, run by the Baker laboratory at the University of Washington. Rosetta@home aims to predict protein–protein docking and design new proteins with the help of about sixty thousand active volunteered computers processing at 83 teraFLOPS on average as of April 18, 2014. Foldit, a Rosetta@Home videogame, aims to reach these goals with a crowdsourcing approach. Though much of the project is oriented towards basic research on improving the accuracy and robustness of the proteomics methods, Rosetta@home also does applied research on malaria, Alzheimer's disease and other pathologies.Like all BOINC projects, Rosetta@home uses idle computer processing resources from volunteers' computers to perform calculations on individual workunits. Completed results are sent to a central project server where they are validated and assimilated into project databases. The project is cross-platform, and runs on a wide variety of hardware configurations. Users can view the progress of their individual protein structure prediction on the Rosetta@home screensaver.In addition to disease-related research, the Rosetta@home network serves as a testing framework for new methods in structural bioinformatics. These new methods are then used in other Rosetta-based applications, like RosettaDock and the Human Proteome Folding Project, after being sufficiently developed and proven stable on Rosetta@home's large and diverse collection of volunteer computers. Two particularly important tests for the new methods developed in Rosetta@home are the Critical Assessment of Techniques for Protein Structure Prediction (CASP) and Critical Assessment of Prediction of Interactions (CAPRI) experiments, biannual experiments which evaluate the state of the art in protein structure prediction and protein–protein docking prediction, respectively. Rosetta@home consistently ranks among the foremost docking predictors, and is one of the best tertiary structure predictors available.