Using Computers to teach Undergraduates about Biological Molecules
... approaches. Students can normally determine the sequence of a polypeptide of 70-80 residues in about two hours. Reasonably realistic yields and 'carry over' contaminations make this an attractive program despite its age. The utility of prediction methods in teaching protein In addition, the structur ...
... approaches. Students can normally determine the sequence of a polypeptide of 70-80 residues in about two hours. Reasonably realistic yields and 'carry over' contaminations make this an attractive program despite its age. The utility of prediction methods in teaching protein In addition, the structur ...
tutorial10_3D_structure
... Protein Data Bank (PDB) • Contains all known 3D structural data of large biological molecules, mostly proteins and nucleic acids: ~87,000 structures. • The data is typically obtained by X-ray crystallography or NMR (Nuclear magnetic resonance) spectroscopy and submitted by biologists and biochemist ...
... Protein Data Bank (PDB) • Contains all known 3D structural data of large biological molecules, mostly proteins and nucleic acids: ~87,000 structures. • The data is typically obtained by X-ray crystallography or NMR (Nuclear magnetic resonance) spectroscopy and submitted by biologists and biochemist ...
Beta-Sheet Structure Prediction Methods
... buried core are critical determinants of the fold. These analyses indicated strong statistical preferences for certain amino acids residues in the folded beta-structural motifs. BETAWRAP predicts beta-helix by dynamically assessing an amino acid segment into stacking beta-strands separated by variab ...
... buried core are critical determinants of the fold. These analyses indicated strong statistical preferences for certain amino acids residues in the folded beta-structural motifs. BETAWRAP predicts beta-helix by dynamically assessing an amino acid segment into stacking beta-strands separated by variab ...
What Are the Best Food Sources of Protein
... d. A 1,600 calorie diet based on MyPlate will supply the protein needs for adult women and most adult men Nutrition & the Human Body-Ch 6 Protein… ...
... d. A 1,600 calorie diet based on MyPlate will supply the protein needs for adult women and most adult men Nutrition & the Human Body-Ch 6 Protein… ...
Biomolecules in water and water in biomolecules
... It is a common understanding that the molecular recognition is an essential elementary process for protein to function. The molecular recognition is a thermodynamic process which is characterized by the free energy difference between two states of a host-guest system, bound and unbound. On the other ...
... It is a common understanding that the molecular recognition is an essential elementary process for protein to function. The molecular recognition is a thermodynamic process which is characterized by the free energy difference between two states of a host-guest system, bound and unbound. On the other ...
051507
... Ch.3 (and lab) • Protein purification – Exploit differences in physical/chemical characteristics (arising from…?) to separate proteins – Ion exchange – Gel filtration/Size exclusion – Affinity ...
... Ch.3 (and lab) • Protein purification – Exploit differences in physical/chemical characteristics (arising from…?) to separate proteins – Ion exchange – Gel filtration/Size exclusion – Affinity ...
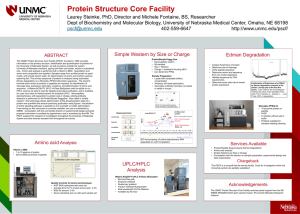
PSCF Poster
... Simple Westerns by Size or Charge Consultation with the director on sample preparation, experimental design and ...
... Simple Westerns by Size or Charge Consultation with the director on sample preparation, experimental design and ...
Align sequence to structure - Computational Bioscience Program
... • Threading: Align sequence to structure (templates) For each alignment, the probability that that each amino acid residue would occur in such an environment is calculated based on observed preferences in determined structures. § Rationale: • Limited number of basic folds found in nature • Amino aci ...
... • Threading: Align sequence to structure (templates) For each alignment, the probability that that each amino acid residue would occur in such an environment is calculated based on observed preferences in determined structures. § Rationale: • Limited number of basic folds found in nature • Amino aci ...
DIAGNOSTIC RELEVANCE OF PREDICTED ANTIGENIC
... Objectives. The purpose of this study was to determinate and evaluate diagnostic relevance of antigenic epitopes encoded by open reading frame for severe acute respiratory syndromecoronavirus (SARS-CoV) nucleocapsid (N) protein. Methods. Three potential antigenic epitopes of SARS-CoV N protein have ...
... Objectives. The purpose of this study was to determinate and evaluate diagnostic relevance of antigenic epitopes encoded by open reading frame for severe acute respiratory syndromecoronavirus (SARS-CoV) nucleocapsid (N) protein. Methods. Three potential antigenic epitopes of SARS-CoV N protein have ...
Student Misconceptions
... molecules are accurate. However, organic molecules are less static than students imagine. Conveniently drawn as linear, monosaccharides usually form rings in aqueous solutions. There may be considerable rotation around single bonds within organic molecules, unless their structure is stabilized by in ...
... molecules are accurate. However, organic molecules are less static than students imagine. Conveniently drawn as linear, monosaccharides usually form rings in aqueous solutions. There may be considerable rotation around single bonds within organic molecules, unless their structure is stabilized by in ...
We propose a frequent pattern-based algorithm for predicting
... Abstract: We propose a frequent pattern-based algorithm for predicting functions and localizations of proteins from their primary structure (amino acid sequence). We use reduced alphabets that capture the higher rate of substitution between amino acids that are physiochemically similar. Frequent sub ...
... Abstract: We propose a frequent pattern-based algorithm for predicting functions and localizations of proteins from their primary structure (amino acid sequence). We use reduced alphabets that capture the higher rate of substitution between amino acids that are physiochemically similar. Frequent sub ...
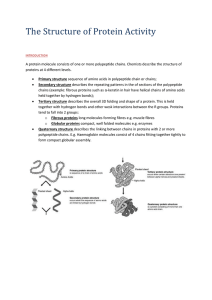
The_Structure_of_Protein_Activity
... b) Which of these amino acids have side chains that: i)are non-polar ii) are polar iii) can ionise? ...
... b) Which of these amino acids have side chains that: i)are non-polar ii) are polar iii) can ionise? ...
In general, animal proteins are considered complete proteins. A complete... essential amino acids. Vegetable (plant-based) proteins are considered incomplete proteins...
... In general, animal proteins are considered complete proteins. A complete protein contains all nine essential amino acids. Vegetable (plant-based) proteins are considered incomplete proteins as they are missing one or more of the essential amino acids. All of the essential amino acids need to be pres ...
... In general, animal proteins are considered complete proteins. A complete protein contains all nine essential amino acids. Vegetable (plant-based) proteins are considered incomplete proteins as they are missing one or more of the essential amino acids. All of the essential amino acids need to be pres ...
ECS 189K - UC Davis
... - The protein chain is defined as a polyline joining the central CA atoms of each residue running from the N-terminal to the C-terminal. Coordinates of the CA atoms are available in the PDB file for the protein of interest - The algorithm attempts to smooth the line as long as it can: it it reaches ...
... - The protein chain is defined as a polyline joining the central CA atoms of each residue running from the N-terminal to the C-terminal. Coordinates of the CA atoms are available in the PDB file for the protein of interest - The algorithm attempts to smooth the line as long as it can: it it reaches ...
Protein Folding Questions only
... functional group. - Hydrophilic sidechains have various combinations of ____________. An exception to this observation is: ...
... functional group. - Hydrophilic sidechains have various combinations of ____________. An exception to this observation is: ...
Fast Categorization of Bacteriophage Protein Families using
... SAM (Sequence Alignment and Modeling) tells us that sequences are related, but there are times when the program is incorrect, and just by looking at a picture, we can tell it’s wrong, or ...
... SAM (Sequence Alignment and Modeling) tells us that sequences are related, but there are times when the program is incorrect, and just by looking at a picture, we can tell it’s wrong, or ...
6. 3-D structure of proteins
... amino acid side chains can stabilize or destabilize this structure. Five different kinds of constraints affect the stability of an α helix: 1. the electrostatic repulsion (or attraction) between successive amino acid residues with charged R group. 2. the bulkiness of adjacent R group 3. the interact ...
... amino acid side chains can stabilize or destabilize this structure. Five different kinds of constraints affect the stability of an α helix: 1. the electrostatic repulsion (or attraction) between successive amino acid residues with charged R group. 2. the bulkiness of adjacent R group 3. the interact ...
Drug_desig_vs7
... The workshops give a simple yet realistic picture of how bioinformatics is used to design drug candidates. Hands-on sessions are for cancer (target BRAF and IDO1) and pain (target COX) treatments. Over 200 people attended our pilot workshops. Many of them, even with a very limited knowledge of chemi ...
... The workshops give a simple yet realistic picture of how bioinformatics is used to design drug candidates. Hands-on sessions are for cancer (target BRAF and IDO1) and pain (target COX) treatments. Over 200 people attended our pilot workshops. Many of them, even with a very limited knowledge of chemi ...
Chapter 1 I am - Mrs Smith`s Biology
... I am the class of protein formed by several spiral-shaped polypeptide molecules becoming linked together in parallel by cross-bridges, giving the protein a rope-like structure Amino Acids ...
... I am the class of protein formed by several spiral-shaped polypeptide molecules becoming linked together in parallel by cross-bridges, giving the protein a rope-like structure Amino Acids ...
The Post-Game/Practice Meal
... DON’T FORGET TO HYDRATE! Rehydration is also an important part of recovery after exercise. In general, 16-24 oz of fluid should be consumed for every pound lost during exercise. For some athletes, drinking a sports drink that contains carbohydrates and electrolytes, such as sodium, can be beneficial ...
... DON’T FORGET TO HYDRATE! Rehydration is also an important part of recovery after exercise. In general, 16-24 oz of fluid should be consumed for every pound lost during exercise. For some athletes, drinking a sports drink that contains carbohydrates and electrolytes, such as sodium, can be beneficial ...
From gene to protein 2
... that are required for their activity To be appropriately modified by protein kinases or other proteinmodifying enzymes ...
... that are required for their activity To be appropriately modified by protein kinases or other proteinmodifying enzymes ...
Protein and Enzyme Check for Understanding
... Protein and Enzyme Check for Understanding: 1. What is the monomer of a protein? 2. What is the name of the bond between the amino acids in a protein? 3. Label the following parts: ...
... Protein and Enzyme Check for Understanding: 1. What is the monomer of a protein? 2. What is the name of the bond between the amino acids in a protein? 3. Label the following parts: ...
Corn Gluten Meal - International Feed
... Corn Gluten Meal Corn Gluten Meal (CGM) is a by-product of corn wet milling process and corn starch production. CGM is a protein-rich feed, containing about 65% crude protein (DM), used as a source of protein, energy and pigments for livestock species including fish. It is also valued in pet food fo ...
... Corn Gluten Meal Corn Gluten Meal (CGM) is a by-product of corn wet milling process and corn starch production. CGM is a protein-rich feed, containing about 65% crude protein (DM), used as a source of protein, energy and pigments for livestock species including fish. It is also valued in pet food fo ...
Rosetta@home

Rosetta@home is a distributed computing project for protein structure prediction on the Berkeley Open Infrastructure for Network Computing (BOINC) platform, run by the Baker laboratory at the University of Washington. Rosetta@home aims to predict protein–protein docking and design new proteins with the help of about sixty thousand active volunteered computers processing at 83 teraFLOPS on average as of April 18, 2014. Foldit, a Rosetta@Home videogame, aims to reach these goals with a crowdsourcing approach. Though much of the project is oriented towards basic research on improving the accuracy and robustness of the proteomics methods, Rosetta@home also does applied research on malaria, Alzheimer's disease and other pathologies.Like all BOINC projects, Rosetta@home uses idle computer processing resources from volunteers' computers to perform calculations on individual workunits. Completed results are sent to a central project server where they are validated and assimilated into project databases. The project is cross-platform, and runs on a wide variety of hardware configurations. Users can view the progress of their individual protein structure prediction on the Rosetta@home screensaver.In addition to disease-related research, the Rosetta@home network serves as a testing framework for new methods in structural bioinformatics. These new methods are then used in other Rosetta-based applications, like RosettaDock and the Human Proteome Folding Project, after being sufficiently developed and proven stable on Rosetta@home's large and diverse collection of volunteer computers. Two particularly important tests for the new methods developed in Rosetta@home are the Critical Assessment of Techniques for Protein Structure Prediction (CASP) and Critical Assessment of Prediction of Interactions (CAPRI) experiments, biannual experiments which evaluate the state of the art in protein structure prediction and protein–protein docking prediction, respectively. Rosetta@home consistently ranks among the foremost docking predictors, and is one of the best tertiary structure predictors available.