Proteins - Mr Waring`s Biology Blog
... A protein consists of one or more polypeptide chains folded into a highly specific 3D shape. There are up to four levels of structure in a protein: primary, secondary, tertiary and quaternary. Each of these play an important role in the overall structure and function of the protein. 7 of 29 ...
... A protein consists of one or more polypeptide chains folded into a highly specific 3D shape. There are up to four levels of structure in a protein: primary, secondary, tertiary and quaternary. Each of these play an important role in the overall structure and function of the protein. 7 of 29 ...
Graduate Biochemistry 7.51: The Major Concepts
... As you will see from the syllabus, the lectures in this course are drawn from a wide range of topics in biochemistry. However, nearly all of the science we discuss is based on a discrete number of fundamental concepts that are common to most biochemical approaches. A major goal of this course is to ...
... As you will see from the syllabus, the lectures in this course are drawn from a wide range of topics in biochemistry. However, nearly all of the science we discuss is based on a discrete number of fundamental concepts that are common to most biochemical approaches. A major goal of this course is to ...
Teaching DNA, Proteins, and Protein Synthesis
... The steps in protein synthesis are easier to understand because proteins are taught first. How changes in DNA affect the shape of proteins will be visualized. We also will connect Mendel’s concepts of genes & traits to the LEGO protein molecules produced. ...
... The steps in protein synthesis are easier to understand because proteins are taught first. How changes in DNA affect the shape of proteins will be visualized. We also will connect Mendel’s concepts of genes & traits to the LEGO protein molecules produced. ...
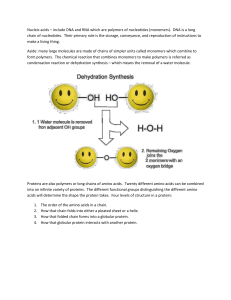
Structure of Proteins
... biological activity are built from the same set of 20 amino acids • Each protein has a distinctive number and specific sequence of amino acid residues • Amino acids are alphabets of protein structure. They can be arranged in an infinite number of sequences to make an infinite number of different pro ...
... biological activity are built from the same set of 20 amino acids • Each protein has a distinctive number and specific sequence of amino acid residues • Amino acids are alphabets of protein structure. They can be arranged in an infinite number of sequences to make an infinite number of different pro ...
Computer science
... transcription factors that bind to DNA near the transcription start site of a gene and influence the rate of transcription. • Goals: identify the transcription factors, characterize the sites they bind to in the genome, and determine how they act in combination to enhance or inhibit transcription. T ...
... transcription factors that bind to DNA near the transcription start site of a gene and influence the rate of transcription. • Goals: identify the transcription factors, characterize the sites they bind to in the genome, and determine how they act in combination to enhance or inhibit transcription. T ...
Protein Structure HW Key
... 16. Discuss how proteins are purified. Depends on the protein, but usually start with some crude source and then a centrifugation step to remove debris. After that, a couple of chromatography steps to purify. 17. What is specific activity? Briefly describe how it is determined. Activity/mg protein. ...
... 16. Discuss how proteins are purified. Depends on the protein, but usually start with some crude source and then a centrifugation step to remove debris. After that, a couple of chromatography steps to purify. 17. What is specific activity? Briefly describe how it is determined. Activity/mg protein. ...
Simian immunodeficiency virus (SIV) (isolate 216.94.A2) gp120
... (isolate 216.94.A2) gp120 Protein (His Tag) Catalog Number: ...
... (isolate 216.94.A2) gp120 Protein (His Tag) Catalog Number: ...
Protein Purification and Analysis Day 4
... at the pH of the running buffer. This charge will, of course, depend on the amino acid composition of the protein as well as post-translational modifications such as addition of sialic acids. Since the protein retains its folded conformation, its hydrodynamic size and mobility on the gel will also v ...
... at the pH of the running buffer. This charge will, of course, depend on the amino acid composition of the protein as well as post-translational modifications such as addition of sialic acids. Since the protein retains its folded conformation, its hydrodynamic size and mobility on the gel will also v ...
Techniques
... _________________________ used for RNA and DNA separation ________________________ gel electrophoresis is used for protein separation ...
... _________________________ used for RNA and DNA separation ________________________ gel electrophoresis is used for protein separation ...
DLS-Characterisation of protein melting point
... melting point Proteins are composed of polypeptide chains, synthesized within the cell from a pool of 20 different amino acid types. In contrast to manmade and random coil biological polymers, the protein’s polypeptide chains are folded into unique 3-dimensional structures in the natured state. Thes ...
... melting point Proteins are composed of polypeptide chains, synthesized within the cell from a pool of 20 different amino acid types. In contrast to manmade and random coil biological polymers, the protein’s polypeptide chains are folded into unique 3-dimensional structures in the natured state. Thes ...
Detecting Protein Function and Protein
... At some point the proteins broke off the same polypeptide, but because of their previous affinity for one another, they now interact. The interfaces between two linked protein domains has been shown to be very similar to that of two separate, interacting proteins. ...
... At some point the proteins broke off the same polypeptide, but because of their previous affinity for one another, they now interact. The interfaces between two linked protein domains has been shown to be very similar to that of two separate, interacting proteins. ...
Document
... How that chain folds into either a pleated sheet or a helix. How that folded chain forms into a globular protein. How that globular protein interacts with another protein. ...
... How that chain folds into either a pleated sheet or a helix. How that folded chain forms into a globular protein. How that globular protein interacts with another protein. ...
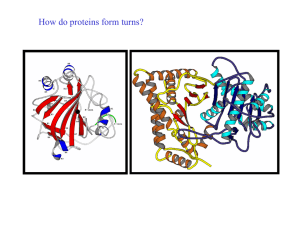
How do proteins form turns? - UF Macromolecular Structure Group
... A reverse turn is region of the polypeptide having a hydrogen bond from one main chain carbonyl oxygen to the main chain N-H group 3 residues along the chain (i.e. O(i) to N(i+3)) Helical regions are excluded from this definition (see later) Reverse turns are very abundant in globular proteins and g ...
... A reverse turn is region of the polypeptide having a hydrogen bond from one main chain carbonyl oxygen to the main chain N-H group 3 residues along the chain (i.e. O(i) to N(i+3)) Helical regions are excluded from this definition (see later) Reverse turns are very abundant in globular proteins and g ...
Structural Genomics - University of Houston
... • Prediction of three dimensional structure from its amino acid sequence ...
... • Prediction of three dimensional structure from its amino acid sequence ...
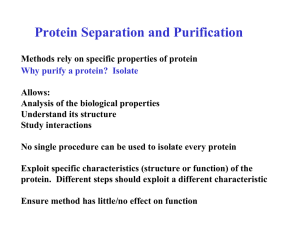
Protein Separation and Purification
... E.g. Antibodies which bind Protein Enzyme which binds a co-enzyme or inhibitor A ligand is covalently bound to a solid matrix (usually agarose) which is then packed into a chromatography column When a mixture containing the protein of interest is applied to the column, the desired protein is bound b ...
... E.g. Antibodies which bind Protein Enzyme which binds a co-enzyme or inhibitor A ligand is covalently bound to a solid matrix (usually agarose) which is then packed into a chromatography column When a mixture containing the protein of interest is applied to the column, the desired protein is bound b ...
Determination of Proteins
... through peptide bonds. Proteins are large molecules and can be split into smaller units by hydrolysis-amino acids. ...
... through peptide bonds. Proteins are large molecules and can be split into smaller units by hydrolysis-amino acids. ...
Quiz Next Tuesday (09/18) - Chemistry at Winthrop University
... second column with different buffer conditions is used to resolve the basic amino acids. (Adapted from Moore, S., Spackman, D., and Stein, ...
... second column with different buffer conditions is used to resolve the basic amino acids. (Adapted from Moore, S., Spackman, D., and Stein, ...
The World of Chemistry Episode 24
... 2. How many subunits are found in hemoglobin? What atom in found in the center of each? There are four subunits, each containing 2 - helices and 2 - sheets. An atom of iron is found in the center of each. 3. Briefly describe the four types of protein structure. Primary - the sequence of amino ac ...
... 2. How many subunits are found in hemoglobin? What atom in found in the center of each? There are four subunits, each containing 2 - helices and 2 - sheets. An atom of iron is found in the center of each. 3. Briefly describe the four types of protein structure. Primary - the sequence of amino ac ...
Episode 24 - The Genetic Code
... 2. How many subunits are found in hemoglobin? What atom in found in the center of each? There are four subunits, each containing 2 - helices and 2 - sheets. An atom of iron is found in the center of each. 3. Briefly describe the four types of protein structure. Primary - the sequence of amino ac ...
... 2. How many subunits are found in hemoglobin? What atom in found in the center of each? There are four subunits, each containing 2 - helices and 2 - sheets. An atom of iron is found in the center of each. 3. Briefly describe the four types of protein structure. Primary - the sequence of amino ac ...
Proteins
... to hydrogen bonds forming between amide and carboxyl groups. There are two possible types of secondary structure: ...
... to hydrogen bonds forming between amide and carboxyl groups. There are two possible types of secondary structure: ...
PHYS-2030 Tutorial 1 1. A protein molecule has a molar mass of
... has a total mass of 40,000 g). The official name for the unit of molar mass is the Dalton (1 Dalton = 1 g/mole, abbreviation = Da). So this protein has a molar mass of 40 kDa. The average density of proteins is about 1300 kg m-3. If the protein molecule is spherical, what is (A) its radius, and (B) ...
... has a total mass of 40,000 g). The official name for the unit of molar mass is the Dalton (1 Dalton = 1 g/mole, abbreviation = Da). So this protein has a molar mass of 40 kDa. The average density of proteins is about 1300 kg m-3. If the protein molecule is spherical, what is (A) its radius, and (B) ...
y-ion series=A, AA, LAA, SLAA
... More likely: 500-1500 embryos Alternative: ES cell differentiation ...
... More likely: 500-1500 embryos Alternative: ES cell differentiation ...
LOYOLA COLLEGE (AUTONOMOUS), CHENNAI – 600 034
... The α and β cyclic forms of D glucose are referred to as__________ The charged amino acid, which is electrically neutral, is called _________. _________ is the specific region on the enzyme at which substrate binds. _________ is the ring system present in cholesterol. Nitrogenous bases are conjugate ...
... The α and β cyclic forms of D glucose are referred to as__________ The charged amino acid, which is electrically neutral, is called _________. _________ is the specific region on the enzyme at which substrate binds. _________ is the ring system present in cholesterol. Nitrogenous bases are conjugate ...
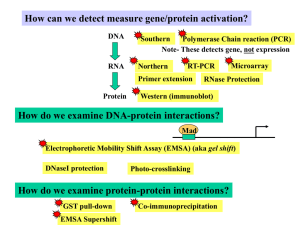
Protein–protein interaction

Protein–protein interactions (PPIs) refer to physical contacts established between two or more proteins as a result of biochemical events and/or electrostatic forces.In fact, proteins are vital macromolecules, at both cellular and systemic levels, but they rarely act alone. Diverse essential molecular processes within a cell are carried out by molecular machines that are built from a large number of protein components organized by their PPIs. Indeed, these interactions are at the core of the entire interactomics system of any living cell and so, unsurprisingly, aberrant PPIs are on the basis of multiple diseases, such as Creutzfeld-Jacob, Alzheimer's disease, and cancer.PPIs have been studied from different perspectives: biochemistry, quantum chemistry, molecular dynamics, signal transduction, among others. All this information enables the creation of large protein interaction networks – similar to metabolic or genetic/epigenetic networks – that empower the current knowledge on biochemical cascades and disease pathogenesis, as well as provide putative new therapeutic targets.