An evaluation of codes more compact than the natural genetic code
... One merit of this proposal from an evolutionary point the natural genetic code could be coded for by the first two of view is the decreased number of possible codes which positions instead of only so few. The justification to propose would result. Here is why this matters. Before a code could a prec ...
... One merit of this proposal from an evolutionary point the natural genetic code could be coded for by the first two of view is the decreased number of possible codes which positions instead of only so few. The justification to propose would result. Here is why this matters. Before a code could a prec ...
View PDF
... encodes for a precise combination of amino acids at the protein level. ATGGATTGCGTG (DNA or gene) changes to AUGGAUUGCGUG (RNA, single gene product) to encode Methionine, Aspartate, Cysteine and Valine amino acid residue, building thereby a crucial motif for the protein function. The motif cannot be ...
... encodes for a precise combination of amino acids at the protein level. ATGGATTGCGTG (DNA or gene) changes to AUGGAUUGCGUG (RNA, single gene product) to encode Methionine, Aspartate, Cysteine and Valine amino acid residue, building thereby a crucial motif for the protein function. The motif cannot be ...
1. Amino Acids,Peptides, Proteins
... Hormones of Pancreas and Gastrointestinal Tract - The photocopy from the 25th edition 23. Thyroid Hormones and Adrenal Medulla Hormones The photocopy from the 25th edition 24. Cholesterol and Bile Acids Ch. 26. Cholesterol Synthesis, Transport, & Excretion - without chemical structures on Figure ...
... Hormones of Pancreas and Gastrointestinal Tract - The photocopy from the 25th edition 23. Thyroid Hormones and Adrenal Medulla Hormones The photocopy from the 25th edition 24. Cholesterol and Bile Acids Ch. 26. Cholesterol Synthesis, Transport, & Excretion - without chemical structures on Figure ...
Nadine Noelting
... Eukaryotic phenylalanine-4-hydroxylase (eu_PheOH); a member of the biopterindependent aromatic amino acid hydroxylase family of non-heme, iron(II)-dependent enzymes that also includes prokaryotic phenylalanine-4-hydroxylase (pro_PheOH), eukaryotic tyrosine hydroxylase (TyrOH) and eukaryotic tryptoph ...
... Eukaryotic phenylalanine-4-hydroxylase (eu_PheOH); a member of the biopterindependent aromatic amino acid hydroxylase family of non-heme, iron(II)-dependent enzymes that also includes prokaryotic phenylalanine-4-hydroxylase (pro_PheOH), eukaryotic tyrosine hydroxylase (TyrOH) and eukaryotic tryptoph ...
Genetics
... Genes are a set of instructions encoded in the DNA sequence of each organism that specify the sequence of amino acids in proteins characteristic of that organism. As a basis for understanding this concept: a. ...
... Genes are a set of instructions encoded in the DNA sequence of each organism that specify the sequence of amino acids in proteins characteristic of that organism. As a basis for understanding this concept: a. ...
DNA_Project - Berkeley Cosmology Group
... from phosphate, a sugar, and one of four nitrogenous bases. The four nitrogenous bases are adenine, thymine, cytosine, and guanine. Based on this cytosine bonds with guanine, and thymine binds with guanine to form bonds between the nucleotides thus creating a strand of DNA. DNA is used in a cell to ...
... from phosphate, a sugar, and one of four nitrogenous bases. The four nitrogenous bases are adenine, thymine, cytosine, and guanine. Based on this cytosine bonds with guanine, and thymine binds with guanine to form bonds between the nucleotides thus creating a strand of DNA. DNA is used in a cell to ...
handout
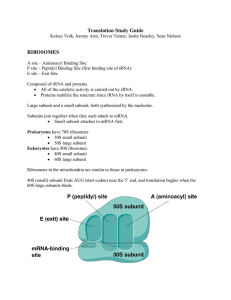
... AUG start codon. 2. Next, the tRNA and small sub-unit complex bind to translation initiation factors and attach to the 5’ cap of the mature mRNA. 3. Next, the entire complex scans in the 3’ direction until it finds the AUG start codon of the mRNA 4. Lastly, the 60S large sub-unit binds to the comple ...
... AUG start codon. 2. Next, the tRNA and small sub-unit complex bind to translation initiation factors and attach to the 5’ cap of the mature mRNA. 3. Next, the entire complex scans in the 3’ direction until it finds the AUG start codon of the mRNA 4. Lastly, the 60S large sub-unit binds to the comple ...
1 Introduction 2 Central Dogma of molecular biology 3 DNA
... the sense that it is also a polymer made up of repeated nucleotides. However, it is single stranded. It is made up of also a different sugar. Its nucleotides are A, U, G, and C, where U is the analog of T in DNA. While most of the RNA gets translated into proteins there are some other types of RNA t ...
... the sense that it is also a polymer made up of repeated nucleotides. However, it is single stranded. It is made up of also a different sugar. Its nucleotides are A, U, G, and C, where U is the analog of T in DNA. While most of the RNA gets translated into proteins there are some other types of RNA t ...
Lecture 9
... evolution. All life evolved from pre-existing life forms. Nature selects among those variants that already exist for the forms best adapted to their particular environments. Many of the ideas that lead to our current view of evolution originated from Charles Darwin and his contemporaries. 1. Chemica ...
... evolution. All life evolved from pre-existing life forms. Nature selects among those variants that already exist for the forms best adapted to their particular environments. Many of the ideas that lead to our current view of evolution originated from Charles Darwin and his contemporaries. 1. Chemica ...
DNA and Central Dogma Study Guide
... 20. Explain transcription in three steps. You should use the terms DNA, RNA polymerase, gene, mRNA, complementary base pairing. a) b) c) 21. What does translation make? 22. Where does translation take place? 23. Explain translation in three steps. You should use the terms ribosomes, codon, amino aci ...
... 20. Explain transcription in three steps. You should use the terms DNA, RNA polymerase, gene, mRNA, complementary base pairing. a) b) c) 21. What does translation make? 22. Where does translation take place? 23. Explain translation in three steps. You should use the terms ribosomes, codon, amino aci ...
3.the nature of proteins
... called amino acids Every amino acid possesses an amino end and a carboxylic acid end There are twenty different naturally occurring amino acids Amino acids differ by virtue of the nature of their R groups Amino acids bond together forming peptide bonds When two amino acids bond during a co ...
... called amino acids Every amino acid possesses an amino end and a carboxylic acid end There are twenty different naturally occurring amino acids Amino acids differ by virtue of the nature of their R groups Amino acids bond together forming peptide bonds When two amino acids bond during a co ...
Chapter 30: Final Questions
... a chemical group for such an interaction. These amino acids might be those for which you would look in your enzyme structure. C). Denote which residue in your enzyme that you might change by site-directed mutagenesis to investigate the fidelity of this enzyme. Which kinetic constant would you expect ...
... a chemical group for such an interaction. These amino acids might be those for which you would look in your enzyme structure. C). Denote which residue in your enzyme that you might change by site-directed mutagenesis to investigate the fidelity of this enzyme. Which kinetic constant would you expect ...
Macromolecules PPT.
... e.g. enzymes – certain enzymes fit certain chemical reactants - heat and change in pH can change the shape of proteins = denaturation - this makes them unable to perform their original functions ...
... e.g. enzymes – certain enzymes fit certain chemical reactants - heat and change in pH can change the shape of proteins = denaturation - this makes them unable to perform their original functions ...
Mutations Learning goals Mutation Where Mutations Occur
... Learning goals • 1. Explain what a mutation is and how it can affect an organism. • 2. Name the two types of cells where mutations can occur and the affects. • 3. Describe the two types of gene mutations and give examples of each. ...
... Learning goals • 1. Explain what a mutation is and how it can affect an organism. • 2. Name the two types of cells where mutations can occur and the affects. • 3. Describe the two types of gene mutations and give examples of each. ...
From Gene to Protein—Transcription and Translation
... In translation, each set of three nucleotides in an mRNA molecule codes for one amino acid in a protein. This explains why each set of three nucleotides in the mRNA is called a codon. Each codon specifies a particular amino acid. For example, the first codon shown above, CGU, instructs the ribosome ...
... In translation, each set of three nucleotides in an mRNA molecule codes for one amino acid in a protein. This explains why each set of three nucleotides in the mRNA is called a codon. Each codon specifies a particular amino acid. For example, the first codon shown above, CGU, instructs the ribosome ...
Document
... – What are the structural features of the sequences of the sequences that you are comparing? Globular/membrane protein? – What is the level of sequence identity of the compared sequences? – Does one MDM fit my data better then the others: You can use ModelGenerator or ProtTest to compare models ...
... – What are the structural features of the sequences of the sequences that you are comparing? Globular/membrane protein? – What is the level of sequence identity of the compared sequences? – Does one MDM fit my data better then the others: You can use ModelGenerator or ProtTest to compare models ...
462a Reading and Homework Assignment 3
... (4) Both cis and trans peptide bonds gain about 85 kJ/mol resonance energy when planar (through orbital alignment). Why are cis peptide bonds rarely seen in proteins? Why are cis peptide bonds more common for prolines than for other amino acids? Steric clash limits cis peptide bonds in most amino ...
... (4) Both cis and trans peptide bonds gain about 85 kJ/mol resonance energy when planar (through orbital alignment). Why are cis peptide bonds rarely seen in proteins? Why are cis peptide bonds more common for prolines than for other amino acids? Steric clash limits cis peptide bonds in most amino ...
Lecture 3: Mutations
... amino acids are coded for by several alternative codons), the resulting new codon may still code for the same amino acid. 2. Missense Mutation: A missense mutation is a nucleotide substitution that changes a codon so that it codes for a different amino acid in the protein. This usually results in a ...
... amino acids are coded for by several alternative codons), the resulting new codon may still code for the same amino acid. 2. Missense Mutation: A missense mutation is a nucleotide substitution that changes a codon so that it codes for a different amino acid in the protein. This usually results in a ...
Cross-species Extrapolation of an Adverse Outcome Pathway for Ecdysteroid Receptor Activation
... Sequence Alignment to Predict Across Species Susceptibility (SeqAPASS) ...
... Sequence Alignment to Predict Across Species Susceptibility (SeqAPASS) ...
SAM Teachers Guide - RI
... Each triplet codes for one amino acid (or stop codon) of a protein chain. ...
... Each triplet codes for one amino acid (or stop codon) of a protein chain. ...
6.2 Human Genetic Disorders
... • Down Syndrome – An extra copy of chromosome 21. Some degree of mental retardation.– Caused during meiosis: chromosomes fail to separate properly. ...
... • Down Syndrome – An extra copy of chromosome 21. Some degree of mental retardation.– Caused during meiosis: chromosomes fail to separate properly. ...
Review - hrsbstaff.ednet.ns.ca
... chromosome is moved to a new location. 4. When a new nucleotide changes the codon to produce a terminator prematurely it is known as _________. 5. When a base pair is removed or a base pair is added, it is known as __________ and ___________ respectively. 6. ___________ is when a new nucleotide prod ...
... chromosome is moved to a new location. 4. When a new nucleotide changes the codon to produce a terminator prematurely it is known as _________. 5. When a base pair is removed or a base pair is added, it is known as __________ and ___________ respectively. 6. ___________ is when a new nucleotide prod ...
No Slide Title
... RNA nucleotides in the cell match up with only one side of the “unzipped” DNA each “unzipped’ strands forms a template for a mRNA strand ...
... RNA nucleotides in the cell match up with only one side of the “unzipped” DNA each “unzipped’ strands forms a template for a mRNA strand ...
Slide 1
... 3.16 Nucleic acids are information-rich polymers of nucleotides A nucleic acid polymer, a polynucleotide, forms from the nucleotide monomers when the phosphate of one nucleotide bonds to the sugar of the next nucleotide – The result is a repeating sugar-phosphate backbone with protruding nitrogen ...
... 3.16 Nucleic acids are information-rich polymers of nucleotides A nucleic acid polymer, a polynucleotide, forms from the nucleotide monomers when the phosphate of one nucleotide bonds to the sugar of the next nucleotide – The result is a repeating sugar-phosphate backbone with protruding nitrogen ...
Chapters 13-16, Molecular Genetics
... c. three base code (43) = a three base code will produce 64 possibilities, more than enough 2. codon = 3 bases found on mRNA a. 3 “stop” codons b. 1 “start” codon c. third base in the codon is often less specific than the first two d. several codons can code for the same amino acid (degenerate) 3. g ...
... c. three base code (43) = a three base code will produce 64 possibilities, more than enough 2. codon = 3 bases found on mRNA a. 3 “stop” codons b. 1 “start” codon c. third base in the codon is often less specific than the first two d. several codons can code for the same amino acid (degenerate) 3. g ...
Genetic code

The genetic code is the set of rules by which information encoded within genetic material (DNA or mRNA sequences) is translated into proteins by living cells. Biological decoding is accomplished by the ribosome, which links amino acids in an order specified by mRNA, using transfer RNA (tRNA) molecules to carry amino acids and to read the mRNA three nucleotides at a time. The genetic code is highly similar among all organisms and can be expressed in a simple table with 64 entries.The code defines how sequences of these nucleotide triplets, called codons, specify which amino acid will be added next during protein synthesis. With some exceptions, a three-nucleotide codon in a nucleic acid sequence specifies a single amino acid. Because the vast majority of genes are encoded with exactly the same code (see the RNA codon table), this particular code is often referred to as the canonical or standard genetic code, or simply the genetic code, though in fact some variant codes have evolved. For example, protein synthesis in human mitochondria relies on a genetic code that differs from the standard genetic code.While the genetic code determines the protein sequence for a given coding region, other genomic regions can influence when and where these proteins are produced.