Evolution, 2e
... Figure 2.10 Evidence for phylogenetic relationships among primates, based on the ψη-globin pseudogene ...
... Figure 2.10 Evidence for phylogenetic relationships among primates, based on the ψη-globin pseudogene ...
Shark Fin Forensics
... then right-click (or ctrl-click) "paste" to paste the other sequence below the great white's sequence. Repeat this process with the other four unknown fins’ 12S sequences until all five unknown sequences are pasted into the same page as the great white 12S sequence. Compare sequences. Compare each ...
... then right-click (or ctrl-click) "paste" to paste the other sequence below the great white's sequence. Repeat this process with the other four unknown fins’ 12S sequences until all five unknown sequences are pasted into the same page as the great white 12S sequence. Compare sequences. Compare each ...
Structural
... The three dimensional folding of a polypeptide is its tertiary structure. Both the a-helix and b-sheet may exist within the tertiary structure. Generally the distribution of amino acid sidechains in a globular protein finds mostly nonpolar residues in the interior of the protein and polar residues o ...
... The three dimensional folding of a polypeptide is its tertiary structure. Both the a-helix and b-sheet may exist within the tertiary structure. Generally the distribution of amino acid sidechains in a globular protein finds mostly nonpolar residues in the interior of the protein and polar residues o ...
MB206_fhs_lnt_001.1_AT_May09
... Double helix is stabilized by H bonds between purine & pyrimidine bases on the opposite strands. A pairs T by 2 H bonds; G pairs C by 3 H bonds. Two strands in DNA helix are complementary, ie. dsDNA contains equimolar amounts of purines (A + G) and pyrimidines (T + C), with A = T and G = C. The ...
... Double helix is stabilized by H bonds between purine & pyrimidine bases on the opposite strands. A pairs T by 2 H bonds; G pairs C by 3 H bonds. Two strands in DNA helix are complementary, ie. dsDNA contains equimolar amounts of purines (A + G) and pyrimidines (T + C), with A = T and G = C. The ...
Molecular Genetic Study of PTC Tasting in Basra
... some genetic parameters of the studied population. Mutation analyzed by Mutation Surveyor Software V. 5. 2. 3. Results and Discussion 3. 1. Gene sequence TAS2R38 P49A The genetic sequence was determined for the PCR product to DNA area consisting of 221bp in the rs713598 region of the gene TAS2R38 (p ...
... some genetic parameters of the studied population. Mutation analyzed by Mutation Surveyor Software V. 5. 2. 3. Results and Discussion 3. 1. Gene sequence TAS2R38 P49A The genetic sequence was determined for the PCR product to DNA area consisting of 221bp in the rs713598 region of the gene TAS2R38 (p ...
Mitochondria tutorial
... However, if we scroll down the list, we see that the enzyme EcoRI fulfills all of our needs --it cuts on either side of the gene sequence but not within the gene sequence itself. Also, the enzyme XbaI might fulfill our requirements, although it looks like the righthand cut site is very close to the ...
... However, if we scroll down the list, we see that the enzyme EcoRI fulfills all of our needs --it cuts on either side of the gene sequence but not within the gene sequence itself. Also, the enzyme XbaI might fulfill our requirements, although it looks like the righthand cut site is very close to the ...
Plant collection protocol
... Short manual on how to collect plant material for DNA purposes For each plant collected the following is compulsory: a) Herbarium voucher: At least two specimens of the same plant must be prepared. One will be kept at the University of Johannesburg Herbarium, and the second deposited to a main herba ...
... Short manual on how to collect plant material for DNA purposes For each plant collected the following is compulsory: a) Herbarium voucher: At least two specimens of the same plant must be prepared. One will be kept at the University of Johannesburg Herbarium, and the second deposited to a main herba ...
Cosmid walking and chromosome jumping in the region of PKD1
... polymorphic locus reported by Breuning et al (4, 5) since this polymorphism marks the proximal boundary of the PKD1 region. It was also important to determine therelativeorientation of these two loci with respect to the chromosome and the distal flanking markers so that cloning of the PKD1 region co ...
... polymorphic locus reported by Breuning et al (4, 5) since this polymorphism marks the proximal boundary of the PKD1 region. It was also important to determine therelativeorientation of these two loci with respect to the chromosome and the distal flanking markers so that cloning of the PKD1 region co ...
Site-specific mutagenesis of M13 clones
... Site-specific mutagenesis of M13 clones (5) Site-specific mutagenesis of M13 clones i. Site-specific mutagenesis involves making a predetermined change in the DNA sequence, unlike random mutagenesis, where the change is made by chance. (Fig. 7.15 and Fig. 7.16) ii. The major difficulty for the metho ...
... Site-specific mutagenesis of M13 clones (5) Site-specific mutagenesis of M13 clones i. Site-specific mutagenesis involves making a predetermined change in the DNA sequence, unlike random mutagenesis, where the change is made by chance. (Fig. 7.15 and Fig. 7.16) ii. The major difficulty for the metho ...
national unit specification: general information
... are expressed and the applications of DNA technology. The Unit allows for the integration of practical activities which could be used in other Units. The essential underpinning knowledge and skills gained here will be useful in Units such as Cell Biology and Biochemistry. ...
... are expressed and the applications of DNA technology. The Unit allows for the integration of practical activities which could be used in other Units. The essential underpinning knowledge and skills gained here will be useful in Units such as Cell Biology and Biochemistry. ...
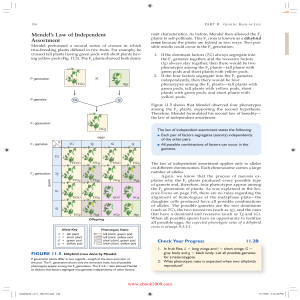
! Mendel`s Law of Independent Assortment
... one shown in Figure 11.5. The Punnett square carries out the multiplication for us, and we add the results to find that the phenotypic ratio is 9:3:3:1. We expect these same results for each and every dihybrid cross. Therefore, it is not necessary to do a Punnett square over and over again for eithe ...
... one shown in Figure 11.5. The Punnett square carries out the multiplication for us, and we add the results to find that the phenotypic ratio is 9:3:3:1. We expect these same results for each and every dihybrid cross. Therefore, it is not necessary to do a Punnett square over and over again for eithe ...
when glucose is scarce
... 1. A repressible operon is one that is usually on; binding of a repressor to the operator shuts off transcription. 1. The trp operon is a repressible operon. trp operon ...
... 1. A repressible operon is one that is usually on; binding of a repressor to the operator shuts off transcription. 1. The trp operon is a repressible operon. trp operon ...
Human Genome Project - the Centre for Applied Genomics
... over five feet, but it is only 50 trillionths of an inch wide. The total amount of dna in the 100 trillion or so cells in the human body laid end to end would run to the sun and back some 20 times. The three billion rungs are made up of chemical units, called “base pairs,” of nucleotides — adenines, ...
... over five feet, but it is only 50 trillionths of an inch wide. The total amount of dna in the 100 trillion or so cells in the human body laid end to end would run to the sun and back some 20 times. The three billion rungs are made up of chemical units, called “base pairs,” of nucleotides — adenines, ...
FLPe Expression Plasmids for E. coli
... 4. Increase the temperature to 37°C and incubate the culture for further 2 3 hours. During this step expression of flpe gene is induced. The FLPe recombinase will subsequently recognise the two aligned FRT sites and all DNA in between (including the selectable marker) gets excised. At the same time ...
... 4. Increase the temperature to 37°C and incubate the culture for further 2 3 hours. During this step expression of flpe gene is induced. The FLPe recombinase will subsequently recognise the two aligned FRT sites and all DNA in between (including the selectable marker) gets excised. At the same time ...
•Fiber diffraction is a method used to determine the structural
... backbones were found to be in the outside of the helix and not inside as it was previously thought to be. They came to this conclusion because of the A and B forms of DNA. The hydrated and dry forms of DNA showed that water could easily come in and bind to DNA, a fact that could only happen if the f ...
... backbones were found to be in the outside of the helix and not inside as it was previously thought to be. They came to this conclusion because of the A and B forms of DNA. The hydrated and dry forms of DNA showed that water could easily come in and bind to DNA, a fact that could only happen if the f ...
Peptide bond Polypeptide
... variable group R is another H atom. But there are 20 different amino acids which are naturally-occurring, and they all have different structures around the R group. There are other amino acids (in fact, thousands more) but these have all been manufactured artificially, and only those 20 occur natura ...
... variable group R is another H atom. But there are 20 different amino acids which are naturally-occurring, and they all have different structures around the R group. There are other amino acids (in fact, thousands more) but these have all been manufactured artificially, and only those 20 occur natura ...
Genetic backgrounds of each Escherichia coli strain used
... galE15: galE mutations are associated with high competence, increased resistance to phage P1 infection, and 2deoxygalactose resistance. galE mutations block the production of UDP-galactose, resulting in truncation of LPS glycans to the minimal, "inner core". The exceptional competence of DH10B is th ...
... galE15: galE mutations are associated with high competence, increased resistance to phage P1 infection, and 2deoxygalactose resistance. galE mutations block the production of UDP-galactose, resulting in truncation of LPS glycans to the minimal, "inner core". The exceptional competence of DH10B is th ...
Mutations, the molecular clock, and models of sequence evolution
... homologous sequences differ at a given position p = 1 – prob. that they are identical p = 1 – (prob. of both staying the same + prob. of both changing to the same thing) p = 1 – { (PAA(t))2 + (PAT(t))2 + (PAC(t))2 + (PAG(t))2 } p = 3/4(1- e-8!t) ...
... homologous sequences differ at a given position p = 1 – prob. that they are identical p = 1 – (prob. of both staying the same + prob. of both changing to the same thing) p = 1 – { (PAA(t))2 + (PAT(t))2 + (PAC(t))2 + (PAG(t))2 } p = 3/4(1- e-8!t) ...
Streptococcus Pyogenes Real Time PCR Kit User Manual
... quencher dye only when the probe hybridizes to the target DNA. This cleavage results in the fluorescent signal generated by the cleaved reporter dye, which is monitored real-time by the PCR detection system. The PCR cycle at which an increase in the fluorescence signal is detected initially (Ct) is ...
... quencher dye only when the probe hybridizes to the target DNA. This cleavage results in the fluorescent signal generated by the cleaved reporter dye, which is monitored real-time by the PCR detection system. The PCR cycle at which an increase in the fluorescence signal is detected initially (Ct) is ...
Chapter 12: Mechanisms and Regulation of Transcription I
... c. RNAP III (PolIII) which transcribes tRNA, snRNA, snoRNA genes as well as the 5S rRNA 5. In structure, the bacterial RNA polymerase most resembles RNA polymerase II from eukaryotes B. RNA Polymerases: The Structure of The Enzyme That Catalyzes Transcription 1. Although RNA polymerases are composed ...
... c. RNAP III (PolIII) which transcribes tRNA, snRNA, snoRNA genes as well as the 5S rRNA 5. In structure, the bacterial RNA polymerase most resembles RNA polymerase II from eukaryotes B. RNA Polymerases: The Structure of The Enzyme That Catalyzes Transcription 1. Although RNA polymerases are composed ...
Nucleic acid analogue
Nucleic acid analogues are compounds which are analogous (structurally similar) to naturally occurring RNA and DNA, used in medicine and in molecular biology research.Nucleic acids are chains of nucleotides, which are composed of three parts: a phosphate backbone, a pucker-shaped pentose sugar, either ribose or deoxyribose, and one of four nucleobases.An analogue may have any of these altered. Typically the analogue nucleobases confer, among other things, different base pairing and base stacking properties. Examples include universal bases, which can pair with all four canonical bases, and phosphate-sugar backbone analogues such as PNA, which affect the properties of the chain (PNA can even form a triple helix).Nucleic acid analogues are also called Xeno Nucleic Acid and represent one of the main pillars of xenobiology, the design of new-to-nature forms of life based on alternative biochemistries.Artificial nucleic acids include peptide nucleic acid (PNA), Morpholino and locked nucleic acid (LNA), as well as glycol nucleic acid (GNA) and threose nucleic acid (TNA). Each of these is distinguished from naturally occurring DNA or RNA by changes to the backbone of the molecule.In May 2014, researchers announced that they had successfully introduced two new artificial nucleotides into bacterial DNA, and by including individual artificial nucleotides in the culture media, were able to passage the bacteria 24 times; they did not create mRNA or proteins able to use the artificial nucleotides. The artificial nucleotides featured 2 fused aromatic rings.