RNA AND PROTEIN SYNTHESIS
... RNA polymerase - the enzyme responsible for RNA transcription. Moves along gene and bonds appropriate RNA nucleotide to complimentary DNA nucleotide. Promoter - binding site on gene that RNA polymerase attaches to at the start of transcription. Codon - set of three mRNA nucleotides that code for an ...
... RNA polymerase - the enzyme responsible for RNA transcription. Moves along gene and bonds appropriate RNA nucleotide to complimentary DNA nucleotide. Promoter - binding site on gene that RNA polymerase attaches to at the start of transcription. Codon - set of three mRNA nucleotides that code for an ...
Protein Complexes – Challenges and Opportunities for
... Resolving the challenge of complexity It is estimated that up to 250.000 protein products are encoded by our genome, and even if only a minor portion is expressed at relevant levels in any type of cell, the number of potential interactions and assemblies is beyond imagination. Thus, their systematic ...
... Resolving the challenge of complexity It is estimated that up to 250.000 protein products are encoded by our genome, and even if only a minor portion is expressed at relevant levels in any type of cell, the number of potential interactions and assemblies is beyond imagination. Thus, their systematic ...
class 1 discussion
... incomplete and computational simulations are so time consuming that prediction of protein function based on only a single DNA sequence is at present impossible (at least for a protein of reasonable size). ...
... incomplete and computational simulations are so time consuming that prediction of protein function based on only a single DNA sequence is at present impossible (at least for a protein of reasonable size). ...
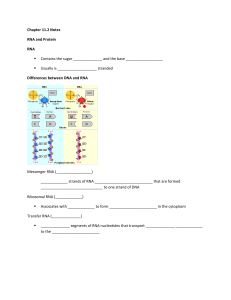
Chapter 11.2 Notes RNA and Protein RNA Contains the sugar and
... ____________________ – the process of ________________________ the info in a sequence of nitrogenous ______________ in mRNA into a sequence of amino acids in _______________ ...
... ____________________ – the process of ________________________ the info in a sequence of nitrogenous ______________ in mRNA into a sequence of amino acids in _______________ ...
FUNCTIONS OF CELL ORGANELLES
... These proteins are called histones. There are five classes of histones- H1,H2A, H2B, H3, H4.These proteins are positively charged and they interact with negatively charged DNA. Two molecules each of H2A, H2B, H3 and H4 form the structural core of the nucleosome.Around this core the segment of DN ...
... These proteins are called histones. There are five classes of histones- H1,H2A, H2B, H3, H4.These proteins are positively charged and they interact with negatively charged DNA. Two molecules each of H2A, H2B, H3 and H4 form the structural core of the nucleosome.Around this core the segment of DN ...
AP Biology - Membrane Structure
... Animal cells need isotonic environment If not, cells must adapt for ...
... Animal cells need isotonic environment If not, cells must adapt for ...
question #5
... Proteins always contain nitrogen because proteins are made of amino acids and amino acids have nitrogen. Nucleic acids always contain nitrogen because nucleic acids consist of three parts, one of which is a nitrogen base. The other two parts are a sugar, which is made up of carbon, hydrogen, and oxy ...
... Proteins always contain nitrogen because proteins are made of amino acids and amino acids have nitrogen. Nucleic acids always contain nitrogen because nucleic acids consist of three parts, one of which is a nitrogen base. The other two parts are a sugar, which is made up of carbon, hydrogen, and oxy ...
- ITA Heidelberg
... interaction. In a fourth type of structure, several protein subunits combine into a complete three-dimensional protein. For example, hemoglobin consists of four subunits. Carbohydrates and lipids are two other basic building blocks of living organisms. Among the carbohydrates, cellulose serves as a ...
... interaction. In a fourth type of structure, several protein subunits combine into a complete three-dimensional protein. For example, hemoglobin consists of four subunits. Carbohydrates and lipids are two other basic building blocks of living organisms. Among the carbohydrates, cellulose serves as a ...
DNA
... • Why: DNA can’t leave the nucleus but the message must get to the ribosome • You are now using U’s no T’s. • RNA polymerase – Enzyme that brings in RNA nucleotides to match up with DNA ...
... • Why: DNA can’t leave the nucleus but the message must get to the ribosome • You are now using U’s no T’s. • RNA polymerase – Enzyme that brings in RNA nucleotides to match up with DNA ...
Slide 1
... • some are attached to inner or outer surface of membrane – peripheral proteins • others embedded in the membrane and span membrane – integral proteins •integral proteins often have carbohydrates attached to them on the exterior face of the cell – these function as receptors, or for cellular recogni ...
... • some are attached to inner or outer surface of membrane – peripheral proteins • others embedded in the membrane and span membrane – integral proteins •integral proteins often have carbohydrates attached to them on the exterior face of the cell – these function as receptors, or for cellular recogni ...
PROTEOME:
... the ions collide with a gas (He, Ne, Ar) resulting in fragmentation of the ion • Fragmentation of the peptides in the collision cell occur in a predictable fashion, mainly at the ...
... the ions collide with a gas (He, Ne, Ar) resulting in fragmentation of the ion • Fragmentation of the peptides in the collision cell occur in a predictable fashion, mainly at the ...
Proteinler - mustafaaltinisik.org.uk
... cleaves at COOH end of Lys and Arg cleaves at COOH end of Phe, Tyr, Trp ...
... cleaves at COOH end of Lys and Arg cleaves at COOH end of Phe, Tyr, Trp ...
Protein Structure and Bioinformatics
... • What are the primary secondary structures? • How are protein structures determined experimentally? • How can structures be predicted in silico? ...
... • What are the primary secondary structures? • How are protein structures determined experimentally? • How can structures be predicted in silico? ...
11046_2011_9445_MOESM6_ESM
... translation, progression of cell cycle, amino acid metabolism, lipid/fatty acid/sterol metabolism and signal transduction showing p-value <0.05, marked with star (*) were considered to be significantly over-represented after hypergeometric probability analysis (www.stattrek.com/tables/hypergeometric ...
... translation, progression of cell cycle, amino acid metabolism, lipid/fatty acid/sterol metabolism and signal transduction showing p-value <0.05, marked with star (*) were considered to be significantly over-represented after hypergeometric probability analysis (www.stattrek.com/tables/hypergeometric ...
Summary for Chapter 6 – Protein: Amino Acids
... Chemically speaking, proteins are more complex than carbohydrates or lipids, being made of some 20 different amino acids, 9 of which the body cannot make; they are essential. Each amino acid contains an amino group, an acid group, a hydrogen atom, and a distinctive side group, all attached to a cent ...
... Chemically speaking, proteins are more complex than carbohydrates or lipids, being made of some 20 different amino acids, 9 of which the body cannot make; they are essential. Each amino acid contains an amino group, an acid group, a hydrogen atom, and a distinctive side group, all attached to a cent ...
any molecule that is present in living organisms. Carbohydrates
... No living organism can Function WITHOUT Enzymes Enzymes: ...
... No living organism can Function WITHOUT Enzymes Enzymes: ...
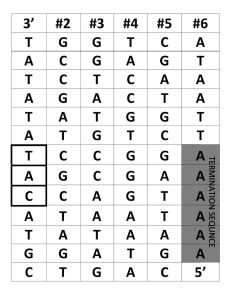
Steps of Translation
... Why is it called translation? • RNA is written in the language of nucleotides – AUG CUA ...
... Why is it called translation? • RNA is written in the language of nucleotides – AUG CUA ...
Transcription
... they are eventually expressed, usually by being translated into amino acid sequences. You may find it helpful to think of exons as DNA that exits the nucleus. In making a primary transcript from a gene, RNA polymerase transcribes both introns and exons from the DNA, but the mRNA molecule that ent ...
... they are eventually expressed, usually by being translated into amino acid sequences. You may find it helpful to think of exons as DNA that exits the nucleus. In making a primary transcript from a gene, RNA polymerase transcribes both introns and exons from the DNA, but the mRNA molecule that ent ...
Describe in simple terms the chemical nature of sugars, proteins
... Sugars – a simple sugar, known as a monosaccharide, is made up of 3 to 7 carbon atoms arranged in a ring. A disaccharide is two monosaccharides, such as glucose and fructose equals sucrose. A polysaccharide is a long chain of monosaccharides and can be either straight or highly branched. Proteins – ...
... Sugars – a simple sugar, known as a monosaccharide, is made up of 3 to 7 carbon atoms arranged in a ring. A disaccharide is two monosaccharides, such as glucose and fructose equals sucrose. A polysaccharide is a long chain of monosaccharides and can be either straight or highly branched. Proteins – ...
Why teach a course in bioinformatics?
... • The simplest way is too search for an open reading frame (ORF). • An ORF is a sequence of codons in DNA that starts with a Start codon, ends with a Stop codon, and has no other Stop codons inside. ...
... • The simplest way is too search for an open reading frame (ORF). • An ORF is a sequence of codons in DNA that starts with a Start codon, ends with a Stop codon, and has no other Stop codons inside. ...
Proposta di ricerca: Introduction Ever since the observation that
... therefore called chaotropes. According to another branch of interpretations, dispersion forces had been suspected to be the main factor responsible for the effects [5]. In our recent work we demonstrated that a unified formalism based on solute-water interfacial tension is able to account for the en ...
... therefore called chaotropes. According to another branch of interpretations, dispersion forces had been suspected to be the main factor responsible for the effects [5]. In our recent work we demonstrated that a unified formalism based on solute-water interfacial tension is able to account for the en ...
shroff srrotary institute of chemical technology
... be obtained by multiplying its nitrogen value by a factor ...
... be obtained by multiplying its nitrogen value by a factor ...
6. protein folding
... code, and as the three-dimensional code, as opposed to the one-dimensional code involved in nucleotide/amino acid sequence. • Importance: – Predict 3D structure from primary sequence – Avoid misfolding related to human diseases – Design proteins with novel functions ...
... code, and as the three-dimensional code, as opposed to the one-dimensional code involved in nucleotide/amino acid sequence. • Importance: – Predict 3D structure from primary sequence – Avoid misfolding related to human diseases – Design proteins with novel functions ...
SR protein

SR proteins are a conserved family of proteins involved in RNA splicing. SR proteins are named because they contain a protein domain with long repeats of serine and arginine amino acid residues, whose standard abbreviations are ""S"" and ""R"" respectively. SR proteins are 50-300 amino acids in length and composed of two domains, the RNA recognition motif (RRM) region and the RS binding domain. SR proteins are more commonly found in the nucleus than the cytoplasm, but several SR proteins are known to shuttle between the nucleus and the cytoplasm.SR proteins were discovered in the 1990s in Drosophila and in amphibian oocytes, and later in humans. In general, metazoans appear to have SR proteins and unicellular organisms lack SR proteins.SR proteins are important in constitutive and alternative pre-mRNA splicing, mRNA export, genome stabilization, nonsense-mediated decay, and translation. SR proteins alternatively splice pre-mRNA by preferentially selecting different splice sites on the pre-mRNA strands to create multiple mRNA transcripts from one pre-mRNA transcript. Once splicing is complete the SR protein may or may not remain attached to help shuttle the mRNA strand out of the nucleus. As RNA Polymerase II is transcribing DNA into RNA, SR proteins attach to newly made pre-mRNA to prevent the pre-mRNA from binding to the coding DNA strand to increase genome stabilization. Topoisomerase I and SR proteins also interact to increase genome stabilization. SR proteins can control the concentrations of specific mRNA that is successfully translated into protein by selecting for nonsense-mediated decay codons during alternative splicing. SR proteins can alternatively splice NMD codons into its own mRNA transcript to auto-regulate the concentration of SR proteins. Through the mTOR pathway and interactions with polyribosomes, SR proteins can increase translation of mRNA.Ataxia telangiectasia, neurofibromatosis type 1, several cancers, HIV-1, and spinal muscular atrophy have all been linked to alternative splicing by SR proteins.