The distribution of DNA translocation times in solid
... electrical driving force = Q ∗ E assuming that Q ∗ does not change as solution viscosity changes. Both Q ∗ E and Fdrag are shown in figure 2(D). This analysis suggests that the estimated electrical driving force exerted on a DNA molecule would be ∼23 pN. The drag force calculated decreased as the so ...
... electrical driving force = Q ∗ E assuming that Q ∗ does not change as solution viscosity changes. Both Q ∗ E and Fdrag are shown in figure 2(D). This analysis suggests that the estimated electrical driving force exerted on a DNA molecule would be ∼23 pN. The drag force calculated decreased as the so ...
A Rapid Chromosome Mapping Method for Cloned Fragments of Yeast DNA.
... containing virtually any yeast gene in which mutants can be found [see BOTSTEIN and DAVIS (1982) for review]. Recombinant DNA methods have, in addition, allowed the identification of interesting DNA segments corresponding to no mapped yeast gene. T h e classical mapping methods referred to can be ap ...
... containing virtually any yeast gene in which mutants can be found [see BOTSTEIN and DAVIS (1982) for review]. Recombinant DNA methods have, in addition, allowed the identification of interesting DNA segments corresponding to no mapped yeast gene. T h e classical mapping methods referred to can be ap ...
Carcinoembryonic Antigens - The Journal of Cell Biology
... set of four alternatively spliced mRNAs which are expressed in several tumor cell lines, in human fetal liver, and in L-cell transfectants. Deduced amino acid sequences of the encoded isoantigens show extensive similarity to CEA and nonspecific cross-reacting antigens, but in addition demonstrate tr ...
... set of four alternatively spliced mRNAs which are expressed in several tumor cell lines, in human fetal liver, and in L-cell transfectants. Deduced amino acid sequences of the encoded isoantigens show extensive similarity to CEA and nonspecific cross-reacting antigens, but in addition demonstrate tr ...
Forensic DNA Fundamentals for the Prosecutor
... in the testing of short tandem repeats (STR). STR testing is a PCR-based technology. As described earlier, genes are specific sequences of nucleotides located at a particular position (locus) on a particular chromosome, and the variant forms of the genes are called alleles.The different alleles are ...
... in the testing of short tandem repeats (STR). STR testing is a PCR-based technology. As described earlier, genes are specific sequences of nucleotides located at a particular position (locus) on a particular chromosome, and the variant forms of the genes are called alleles.The different alleles are ...
DNA Denaturing through UV-C Photon Dissipation: A
... temperature) while the scattering is constant or only slightly increasing with denaturing for the large yeast (figure 1(b)) and salmon sperm DNA (not shown). This result, interpreted as being due to the fact that that complete denaturing occurs for the small synthetic DNA (even at low temperature) w ...
... temperature) while the scattering is constant or only slightly increasing with denaturing for the large yeast (figure 1(b)) and salmon sperm DNA (not shown). This result, interpreted as being due to the fact that that complete denaturing occurs for the small synthetic DNA (even at low temperature) w ...
R4, a non-LTR retrotransposon specific to the
... The specific location of Rl and R2 elements within the 28S rRNA genes of the host simplifies their identification in other species by either Southern blotting or PCR amplification (1,18). PCR is the more sensitive of the two approaches and we are currently able to detect R1 and/or R2 elements in ins ...
... The specific location of Rl and R2 elements within the 28S rRNA genes of the host simplifies their identification in other species by either Southern blotting or PCR amplification (1,18). PCR is the more sensitive of the two approaches and we are currently able to detect R1 and/or R2 elements in ins ...
Replication and Recombinantion
... template for two new strands which then join together, giving two old strands together and two new Dispersive - In strands together which sections of the old strands are dispersed in the new strands ...
... template for two new strands which then join together, giving two old strands together and two new Dispersive - In strands together which sections of the old strands are dispersed in the new strands ...
CIRCULAR DNA MOLECULES IN THE GENUS
... covalently closed circular duplex DNA’s, and their sedimentation coefficients, rather than being less, as is the case with linear molecules, are even greater than they are at neutral pH. We have made only tentative measurements of the sedimentation coefficients of the DNA’s from three of the species ...
... covalently closed circular duplex DNA’s, and their sedimentation coefficients, rather than being less, as is the case with linear molecules, are even greater than they are at neutral pH. We have made only tentative measurements of the sedimentation coefficients of the DNA’s from three of the species ...
video slide - Biology at Mott
... At the end of each replication bubble is a replication fork, a Y-shaped region where new DNA strands are elongating Helicases are enzymes that untwist the double helix at the replication forks Single-strand binding protein binds to and stabilizes single-stranded DNA until it can be used as a templat ...
... At the end of each replication bubble is a replication fork, a Y-shaped region where new DNA strands are elongating Helicases are enzymes that untwist the double helix at the replication forks Single-strand binding protein binds to and stabilizes single-stranded DNA until it can be used as a templat ...
Chapter 21
... An insertion sequence is a transposon that codes for the enzyme(s) needed for transposition flanked by short inverted terminal repeats. The target site at which a transposon is inserted is duplicated during the insertion process to form two repeats in direct orientation at the ends of the transposon ...
... An insertion sequence is a transposon that codes for the enzyme(s) needed for transposition flanked by short inverted terminal repeats. The target site at which a transposon is inserted is duplicated during the insertion process to form two repeats in direct orientation at the ends of the transposon ...
Sterile, 24-well tissue culture plates are filled with melted minimal ... 1.0 ml per well using a repeating syringe. After the...
... digested total, genomic DNA from several species of Phytophthora, including P. cinnamomi, and several species of Boletus, Chroogomphus vinicolor, Gomphidius glutinosus, Leccinum manzanitae, Magnaporthe grisea, Neurospora crassa, N. tetrasperma, Omphalotus olivascens, and Talaromyces flavus. Yield wa ...
... digested total, genomic DNA from several species of Phytophthora, including P. cinnamomi, and several species of Boletus, Chroogomphus vinicolor, Gomphidius glutinosus, Leccinum manzanitae, Magnaporthe grisea, Neurospora crassa, N. tetrasperma, Omphalotus olivascens, and Talaromyces flavus. Yield wa ...
記錄 編號 3862 狀態 NC090FJU00112010 助教 查核 索書 號 學校
... pestis. Further analysis of the region flanking this fragment showed structural features of a bacterial insertion sequence (IS) element. DNA sequence analysis indicated that this IS is 1956bp in length and delimited by two imperfect inverted repeats of 29bp with 8 mismatches. Besides, less conserved ...
... pestis. Further analysis of the region flanking this fragment showed structural features of a bacterial insertion sequence (IS) element. DNA sequence analysis indicated that this IS is 1956bp in length and delimited by two imperfect inverted repeats of 29bp with 8 mismatches. Besides, less conserved ...
groups, blaOXY-1 and blaOXY-2. Klebsiella oxytoca are divided into
... Similarity with other b-lactamases. The percentage similarities were determined for entire proteins. The OXY b-lactamases belong to class A as previously reported (3). The blaOXY group includes blaOXY-1 types such as E23004 (3) or SL781 (16) and blaOXY-2 types such as D488 (38) or SL911. This group ...
... Similarity with other b-lactamases. The percentage similarities were determined for entire proteins. The OXY b-lactamases belong to class A as previously reported (3). The blaOXY group includes blaOXY-1 types such as E23004 (3) or SL781 (16) and blaOXY-2 types such as D488 (38) or SL911. This group ...
Tuning Biphenyl Dioxygenase for Extended Substrate Specificity
... 1986; Bopp, 1986). Due to its broad substrate specificity, it was used as the starting template for generating variant enzymes that can act on an extended set of PCB congeners. Because substrate specificity is determined mainly by the bphA region, only this gene fragment was subjected to directed ev ...
... 1986; Bopp, 1986). Due to its broad substrate specificity, it was used as the starting template for generating variant enzymes that can act on an extended set of PCB congeners. Because substrate specificity is determined mainly by the bphA region, only this gene fragment was subjected to directed ev ...
Molecular cloning of a cDNA encoding a novel Ca2+
... cloned cDNA encodes a nuclease. This is the ¢rst report that a plant nuclease homolog is functional. We further tested e¡ects of the divalent cations, Ca2 , Mg2 , and Zn2 on the activity of the nuclease. The presence of EGTA (grid C2) and EDTA (grid C3) inhibited nuclease activity, as well as DNa ...
... cloned cDNA encodes a nuclease. This is the ¢rst report that a plant nuclease homolog is functional. We further tested e¡ects of the divalent cations, Ca2 , Mg2 , and Zn2 on the activity of the nuclease. The presence of EGTA (grid C2) and EDTA (grid C3) inhibited nuclease activity, as well as DNa ...
DNA structure 2008
... The DNA replicates in interphase beginning at the origins of replication and proceeding bidirectionally from the origins across the chromosome. Many origins are required to ensure that the entire chromosome can be replicated rapidly. In M phase, the centromere attaches the duplicated chromosome to t ...
... The DNA replicates in interphase beginning at the origins of replication and proceeding bidirectionally from the origins across the chromosome. Many origins are required to ensure that the entire chromosome can be replicated rapidly. In M phase, the centromere attaches the duplicated chromosome to t ...
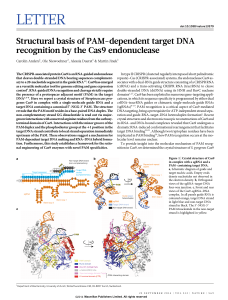
Structural basis of PAM-dependent target DNA recognition by the
... in complex with an 83-nucleotide sgRNA and a partially duplexed target DNA containing a 59-TGG-39 PAM sequence (Fig. 1 and Extended Data Table 1). Owing to an inactivating mutation (H840A) in the Cas9 HNH nuclease domain, the structure reveals an intact target (complementary) DNA strand, while the n ...
... in complex with an 83-nucleotide sgRNA and a partially duplexed target DNA containing a 59-TGG-39 PAM sequence (Fig. 1 and Extended Data Table 1). Owing to an inactivating mutation (H840A) in the Cas9 HNH nuclease domain, the structure reveals an intact target (complementary) DNA strand, while the n ...
PDF
... genes have extensive similarity (BLASTP; 1e-10) to those of B. Thirty-seven families of protein-coding repetitive sehalodurans. Their overall genome similarity ranks the highest quences longer than 300 bp were also categorized. Most of among all the sequenced genomes, regardless if they are therthem ...
... genes have extensive similarity (BLASTP; 1e-10) to those of B. Thirty-seven families of protein-coding repetitive sehalodurans. Their overall genome similarity ranks the highest quences longer than 300 bp were also categorized. Most of among all the sequenced genomes, regardless if they are therthem ...
Functional analysis of plastid DNA replication origins in tobacco by
... structure (Kunnimalaiyaan and Nielsen, 1997b). Interestingly, there are two elements in the near surrounding of this sequence which could also be of importance in plastid DNA replication: (a) The region upstream of the trnI gene shows striking length and sequence differences in the plastid DNA of di ...
... structure (Kunnimalaiyaan and Nielsen, 1997b). Interestingly, there are two elements in the near surrounding of this sequence which could also be of importance in plastid DNA replication: (a) The region upstream of the trnI gene shows striking length and sequence differences in the plastid DNA of di ...
Force spectroscopy of single DNA and RNA molecules Mark C
... toroidal structures [8]. Single-molecule stretching methods have been used to measure the forces that cause this DNA condensation. If the two ends of a DNA molecule are stretched, they are prevented from condensing. The force required to prevent condensation, the attractive condensation force, has b ...
... toroidal structures [8]. Single-molecule stretching methods have been used to measure the forces that cause this DNA condensation. If the two ends of a DNA molecule are stretched, they are prevented from condensing. The force required to prevent condensation, the attractive condensation force, has b ...
Overview-of-CF-and-CF-Genotyping-Platforms
... Was the baby homozygous or hemizygous for Y1092H T>C? • hemizygous is when there is only 1 member of a chromosome segment rather than the usual 2 ...
... Was the baby homozygous or hemizygous for Y1092H T>C? • hemizygous is when there is only 1 member of a chromosome segment rather than the usual 2 ...
Evolutionary Origin and Adaptive Function of Meiosis
... 2004). In eukaryotes such as mammals, tens to hundreds of thousands of naturally caused DNA damages occur per cell per day (see next section). While most of these DNA damages can be repaired, such repair is not 100% efficient. Unrepaired DNA damages accumulate, especially in non-replicating or slowl ...
... 2004). In eukaryotes such as mammals, tens to hundreds of thousands of naturally caused DNA damages occur per cell per day (see next section). While most of these DNA damages can be repaired, such repair is not 100% efficient. Unrepaired DNA damages accumulate, especially in non-replicating or slowl ...
A physical map of the genome of Hmmophilus
... Digestion of DNA in agarose blocks. Usually digests were carried out on the DNA contained in one-third of a complete plug. Restriction einzyme buffers were diffused into the agarose blocks as outlined below. Plugs or portions of plugs were washed in Eppendorf tubes with 500 1.11 vlolumesof buffer (u ...
... Digestion of DNA in agarose blocks. Usually digests were carried out on the DNA contained in one-third of a complete plug. Restriction einzyme buffers were diffused into the agarose blocks as outlined below. Plugs or portions of plugs were washed in Eppendorf tubes with 500 1.11 vlolumesof buffer (u ...
Separating endogenous ancient DNA from modern day
... pled sequences from the 100-y-old remains of an Australian individual (27), four ∼5,000-y-old Neolithic Scandinavian individuals (8), four 38,000- to 70,000-y-old Neandertal individuals (24), and four present day individuals (24). These individuals showed evidence of PMD roughly proportional to thei ...
... pled sequences from the 100-y-old remains of an Australian individual (27), four ∼5,000-y-old Neolithic Scandinavian individuals (8), four 38,000- to 70,000-y-old Neandertal individuals (24), and four present day individuals (24). These individuals showed evidence of PMD roughly proportional to thei ...