A familial inverted duplication/deletion of 2p25.1–25.3
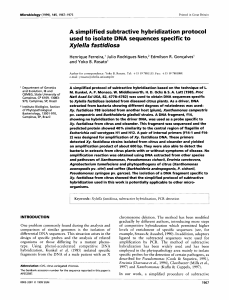
... Figure 1 Overview of molecular data from Case 2. Identical results were obtained on Case 1 and Case 3. (a) Cytogenetics and FISH results. (from left) cut-out of the normal and abnormal chromosomes 2 in G-banding at a resolution of 550 bands. Ideogram of normal and inverted duplicated chromosomes 2p: ...
... Figure 1 Overview of molecular data from Case 2. Identical results were obtained on Case 1 and Case 3. (a) Cytogenetics and FISH results. (from left) cut-out of the normal and abnormal chromosomes 2 in G-banding at a resolution of 550 bands. Ideogram of normal and inverted duplicated chromosomes 2p: ...
document
... and subsequently turning on Trecharge and Tgate, as shown in Fig. 2b. The sliding-time window is applied globally to the entire array. The inpixel 1-bit memory holds the state of the SPAD between readout cycles. When the system is operating at 40MHz, 16,384 excitation laser pulses reach the fluoresc ...
... and subsequently turning on Trecharge and Tgate, as shown in Fig. 2b. The sliding-time window is applied globally to the entire array. The inpixel 1-bit memory holds the state of the SPAD between readout cycles. When the system is operating at 40MHz, 16,384 excitation laser pulses reach the fluoresc ...
DNA Barcoding of Algae and Bacteria from
... the NCBI website was used to analyze the DNA sequences resulting from sequencing. Bioinformatics programs were then used to identify the different species of Algae in different environments based on the BLAST results analysis. DAY 2 ...
... the NCBI website was used to analyze the DNA sequences resulting from sequencing. Bioinformatics programs were then used to identify the different species of Algae in different environments based on the BLAST results analysis. DAY 2 ...
A array
... cRNA quantitation and process control will help. • A hybrid approach based on global scaling and spikes performs the same as global AD scaling for single designs, and also allows cross-design comparisons ...
... cRNA quantitation and process control will help. • A hybrid approach based on global scaling and spikes performs the same as global AD scaling for single designs, and also allows cross-design comparisons ...
File - Molecular Biology 2
... molecular geneticists with methods by which genes or other segments of large chromosomes can be isolated, replicated, and studied by nucleic acid sequencing techniques, electron microscopy, and other analytical techniques. Indeed, genes or other DNA sequences can be amplified by two distinct approac ...
... molecular geneticists with methods by which genes or other segments of large chromosomes can be isolated, replicated, and studied by nucleic acid sequencing techniques, electron microscopy, and other analytical techniques. Indeed, genes or other DNA sequences can be amplified by two distinct approac ...
Electrophoresis, Blotting and Immunodetection Gel
... gel-purified PCR products for sequencing or cloning without need for further purification. Device volume is 100µL of gel. Typically recovers >70% of 100 to 10,000bp DNA from standard agarose gels. Kit components: 50 gel nebulisers; 50 Ultrafree-MC 0.45µm separators; 50 microcentrifuge vials; 500mL s ...
... gel-purified PCR products for sequencing or cloning without need for further purification. Device volume is 100µL of gel. Typically recovers >70% of 100 to 10,000bp DNA from standard agarose gels. Kit components: 50 gel nebulisers; 50 Ultrafree-MC 0.45µm separators; 50 microcentrifuge vials; 500mL s ...
Physicochemical studies on interactions between DNA and RNA
... expected ratio of 4 for the saturation of the 4 promoters on this fragment. The trivial possibility that a large fraction of enzyme preparation is inactive is highly unlikely from results described in the first two papers of this series (Hsieh and Wang, 1976; Wang et al., 1977). ...
... expected ratio of 4 for the saturation of the 4 promoters on this fragment. The trivial possibility that a large fraction of enzyme preparation is inactive is highly unlikely from results described in the first two papers of this series (Hsieh and Wang, 1976; Wang et al., 1977). ...
Chapter 20
... enzyme digestion, is separated into “bands”; each band contains thousands of molecules of the same length. After the current is turned off, a DNA-binding dye is added. This dye fluoresces pink in ultraviolet light, revealing the separated bands to which it binds. In this actual gel, the pink bands c ...
... enzyme digestion, is separated into “bands”; each band contains thousands of molecules of the same length. After the current is turned off, a DNA-binding dye is added. This dye fluoresces pink in ultraviolet light, revealing the separated bands to which it binds. In this actual gel, the pink bands c ...
Genetics, environment and cognitive abilities
... An approach to DNA pooling has been developed as part of our project on g comparing the allele image patterns (AIPs) from an automated DNA sequencer for two pools of DNA (Daniels et al, al, 1998). The DNA pooling method has been shown to be a reasonably accurate screening tool to detect the largest ...
... An approach to DNA pooling has been developed as part of our project on g comparing the allele image patterns (AIPs) from an automated DNA sequencer for two pools of DNA (Daniels et al, al, 1998). The DNA pooling method has been shown to be a reasonably accurate screening tool to detect the largest ...
Objective 2.1 Lesson D Recombinant Organisms
... Show a video clip (appropriate segment begins at 25 minutes) from the series DNA: Playing God, which gives an overview of how insulin was originally harvested from cows and how scientists were able to genetically engineer bacteria to grow insulin in large quantities. The video explains how the gene ...
... Show a video clip (appropriate segment begins at 25 minutes) from the series DNA: Playing God, which gives an overview of how insulin was originally harvested from cows and how scientists were able to genetically engineer bacteria to grow insulin in large quantities. The video explains how the gene ...
Tracking bacterial DNA replication forks in vivo by pulsed field gel
... chromosomal DNA replication with PFG electrophoresis. E. coli strain AQ2, derived from isolate 15, was pulse labeled with 14C-thymidine at various times during amino acid starvation and after the start of synchronous DNA synthesis. Intact chromosomal DNA was prepared in agarose and digested with the ...
... chromosomal DNA replication with PFG electrophoresis. E. coli strain AQ2, derived from isolate 15, was pulse labeled with 14C-thymidine at various times during amino acid starvation and after the start of synchronous DNA synthesis. Intact chromosomal DNA was prepared in agarose and digested with the ...
1-2 Teacher
... Separating DNA In gel electrophoresis, DNA fragments are placed at one end of a porous gel, and an electric voltage is applied to the gel. When the power is turned on, the negativelycharged DNA molecules move toward the positive end of the gel. ...
... Separating DNA In gel electrophoresis, DNA fragments are placed at one end of a porous gel, and an electric voltage is applied to the gel. When the power is turned on, the negativelycharged DNA molecules move toward the positive end of the gel. ...
DATA ENCRYPTION USING BIO MOLECULAR INFORMATION
... Process of converting messages from plain text to cipher text is called cryptography. Cryptography is a technique of achieving security for communications by encoding plain text messages to make it unreadable[1]. Encryption is a useful tool in protecting confidentiality and integrity of information. ...
... Process of converting messages from plain text to cipher text is called cryptography. Cryptography is a technique of achieving security for communications by encoding plain text messages to make it unreadable[1]. Encryption is a useful tool in protecting confidentiality and integrity of information. ...
S1 Text. Supplementary Methods
... calls in the inbred C. rubella line Cr1GR1, which is expected to be highly homozygous. Regions with evidence for high proportions of repeats, copy number variation or high proportion of heterozygous calls in Cr1GR1 were removed from consideration in further analyses of allele-specific expression. To ...
... calls in the inbred C. rubella line Cr1GR1, which is expected to be highly homozygous. Regions with evidence for high proportions of repeats, copy number variation or high proportion of heterozygous calls in Cr1GR1 were removed from consideration in further analyses of allele-specific expression. To ...
Ever since the days of Rene Descartes, the French philosopher
... from the agarose gel and extracted Figure 11.3 A typical agarose gel from the gel piece. This step is known electrophoresis showing migration of undigested as elution. The DNA fragments (lane 1) and digested set of purified in this way are used in DNA fragments (lane 2 to 4) constructing recombinant ...
... from the agarose gel and extracted Figure 11.3 A typical agarose gel from the gel piece. This step is known electrophoresis showing migration of undigested as elution. The DNA fragments (lane 1) and digested set of purified in this way are used in DNA fragments (lane 2 to 4) constructing recombinant ...
Cytogenetics Cytogenetics
... Example • The first chromosome, long arm, second region of the chromosome, the fourth band of that sub-region ...
... Example • The first chromosome, long arm, second region of the chromosome, the fourth band of that sub-region ...
Chapter 8: Variations in Chromosome Number and
... Most members of diploid species contain two haploid sets of chromosomes, but there are variations from this pattern. These changes include a difference in chromosome number, deletion or duplication of genes, and rearrangement of genetic material among or within the chromosomes. All of these change ...
... Most members of diploid species contain two haploid sets of chromosomes, but there are variations from this pattern. These changes include a difference in chromosome number, deletion or duplication of genes, and rearrangement of genetic material among or within the chromosomes. All of these change ...
Document
... Gene therapy aims to treat a disease by supplying a functional allele One possible procedure – Clone the functional allele and insert it in a retroviral vector – Use the virus to deliver the gene to an affected cell type from the patient, such as a bone marrow cell – Viral DNA and the functional ...
... Gene therapy aims to treat a disease by supplying a functional allele One possible procedure – Clone the functional allele and insert it in a retroviral vector – Use the virus to deliver the gene to an affected cell type from the patient, such as a bone marrow cell – Viral DNA and the functional ...
CHROMOSOMES AND DISEASE
... individual (one who possesses two or more genetically different cell lines derived from a single zygote). -Numerical abnormalities: There can be extra copies of the autosomes or the sex chromosomes. Aneuploidy represents an abnormal chromosomal number due to an extra (trisomy) or missing (monosomy) ...
... individual (one who possesses two or more genetically different cell lines derived from a single zygote). -Numerical abnormalities: There can be extra copies of the autosomes or the sex chromosomes. Aneuploidy represents an abnormal chromosomal number due to an extra (trisomy) or missing (monosomy) ...
pcr
... • permits multiple targets to be amplified with only a single primer pair, thus avoiding the resolution limitations of multiplex PCR. • Multiplex-PCR: The use of multiple, unique primer sets within a single PCR reaction to produce amplicons of varying sizes specific to different DNA sequences. • By ...
... • permits multiple targets to be amplified with only a single primer pair, thus avoiding the resolution limitations of multiplex PCR. • Multiplex-PCR: The use of multiple, unique primer sets within a single PCR reaction to produce amplicons of varying sizes specific to different DNA sequences. • By ...
Comparative genomic hybridization

Comparative genomic hybridization is a molecular cytogenetic method for analysing copy number variations (CNVs) relative to ploidy level in the DNA of a test sample compared to a reference sample, without the need for culturing cells. The aim of this technique is to quickly and efficiently compare two genomic DNA samples arising from two sources, which are most often closely related, because it is suspected that they contain differences in terms of either gains or losses of either whole chromosomes or subchromosomal regions (a portion of a whole chromosome). This technique was originally developed for the evaluation of the differences between the chromosomal complements of solid tumor and normal tissue, and has an improved resoIution of 5-10 megabases compared to the more traditional cytogenetic analysis techniques of giemsa banding and fluorescence in situ hybridization (FISH) which are limited by the resolution of the microscope utilized.This is achieved through the use of competitive fluorescence in situ hybridization. In short, this involves the isolation of DNA from the two sources to be compared, most commonly a test and reference source, independent labelling of each DNA sample with a different fluorophores (fluorescent molecules) of different colours (usually red and green), denaturation of the DNA so that it is single stranded, and the hybridization of the two resultant samples in a 1:1 ratio to a normal metaphase spread of chromosomes, to which the labelled DNA samples will bind at their locus of origin. Using a fluorescence microscope and computer software, the differentially coloured fluorescent signals are then compared along the length of each chromosome for identification of chromosomal differences between the two sources. A higher intensity of the test sample colour in a specific region of a chromosome indicates the gain of material of that region in the corresponding source sample, while a higher intensity of the reference sample colour indicates the loss of material in the test sample in that specific region. A neutral colour (yellow when the fluorophore labels are red and green) indicates no difference between the two samples in that location.CGH is only able to detect unbalanced chromosomal abnormalities. This is because balanced chromosomal abnormalities such as reciprocal translocations, inversions or ring chromosomes do not affect copy number, which is what is detected by CGH technologies. CGH does, however, allow for the exploration of all 46 human chromosomes in single test and the discovery of deletions and duplications, even on the microscopic scale which may lead to the identification of candidate genes to be further explored by other cytological techniques.Through the use of DNA microarrays in conjunction with CGH techniques, the more specific form of array CGH (aCGH) has been developed, allowing for a locus-by-locus measure of CNV with increased resolution as low as 100 kilobases. This improved technique allows for the aetiology of known and unknown conditions to be discovered.























