the evolution of genomic base composition in bacteria
... eubacterial species were obtained from the Codon Usage Database (Nakamura et al. 1997; http://www.kazusa.or.jp/ codon/), which provides codon usage and GC content information based on the protein-coding genes available in GenBank (Table 1, Fig. 1). To ensure that the genomic estimates were accurate, ...
... eubacterial species were obtained from the Codon Usage Database (Nakamura et al. 1997; http://www.kazusa.or.jp/ codon/), which provides codon usage and GC content information based on the protein-coding genes available in GenBank (Table 1, Fig. 1). To ensure that the genomic estimates were accurate, ...
Level 3, 2004
... the order, and how many of them are linked together, determines what the protein is and does. These amino acid sequences are coded for by sequenced triplets of bases on the DNA. If the DNA has a large part of the sequence replaced by another sequence, then it usually forms a completely different pro ...
... the order, and how many of them are linked together, determines what the protein is and does. These amino acid sequences are coded for by sequenced triplets of bases on the DNA. If the DNA has a large part of the sequence replaced by another sequence, then it usually forms a completely different pro ...
Restriction Enzyme Digest and Plasmid mapping
... same enzyme is also used to cut the DNA of the recipient into which the fragment will be inserted. Restriction enzymes are proteins that cut DNA at specific sites. Restriction enzymes, also known as restriction endonucleases, recognize specific sequences of DNA base pairs and cut, or chemically sepa ...
... same enzyme is also used to cut the DNA of the recipient into which the fragment will be inserted. Restriction enzymes are proteins that cut DNA at specific sites. Restriction enzymes, also known as restriction endonucleases, recognize specific sequences of DNA base pairs and cut, or chemically sepa ...
Chapter 19
... Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. ...
... Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. ...
Genetic Basis of Polymurphism in the Color Vision of
... least mean difference squared between the data set and the best fitting combination) was 3.1 x low3 log unit for the 534J550 combination and 6.0 x IOs4 log unit for the 5501561 combination. This indicates (Fig. 1) that this monkey has the 55OJ561 pigment combination. The cone pigment phenotypes of t ...
... least mean difference squared between the data set and the best fitting combination) was 3.1 x low3 log unit for the 534J550 combination and 6.0 x IOs4 log unit for the 5501561 combination. This indicates (Fig. 1) that this monkey has the 55OJ561 pigment combination. The cone pigment phenotypes of t ...
ppt - Chair of Computational Biology
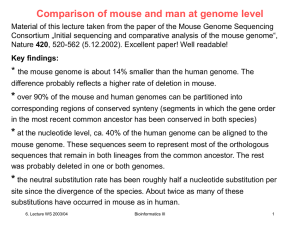
... sequences that remain in both lineages from the common ancestor. The rest was probably deleted in one or both genomes. ...
... sequences that remain in both lineages from the common ancestor. The rest was probably deleted in one or both genomes. ...
The Microbiology of Bad Breath and Periodontitis
... continued… • High VSC concentrations indicate oral malodor • Volatile Sulfur Compounds (VSC) are measured with a ...
... continued… • High VSC concentrations indicate oral malodor • Volatile Sulfur Compounds (VSC) are measured with a ...
Use of QTL analysis in physiological research
... obtained by repeated self-pollination and single seed descent of F2 obtained after crossing two divergent accessions. In this species, an increasing collection of RIL populations is becoming available and has been used to analyze a wide range of traits [1, 2]. The principle of QTL mapping using a RI ...
... obtained by repeated self-pollination and single seed descent of F2 obtained after crossing two divergent accessions. In this species, an increasing collection of RIL populations is becoming available and has been used to analyze a wide range of traits [1, 2]. The principle of QTL mapping using a RI ...
Friends Foes Bacterial Friends and Foes
... were tested for E. coli using aerobic (oxygen-rich) counting techniques, far fewer bacteria would be found, because they recover more slowly under these conditions than if the test were conducted under anaerobic conditions where E.coli would have recovered faster and started to multiply.This could r ...
... were tested for E. coli using aerobic (oxygen-rich) counting techniques, far fewer bacteria would be found, because they recover more slowly under these conditions than if the test were conducted under anaerobic conditions where E.coli would have recovered faster and started to multiply.This could r ...
Navigating the NCBI Intructions
... 12. Click on the BRCA1 link. This will take you to Entrez Gene, which provides a summary of the information available at the NCBI for BRCA1. Scroll through the webpage and explore some of the information available. Scroll down the webpage to the section titled “Gene Ontology.” There is a table title ...
... 12. Click on the BRCA1 link. This will take you to Entrez Gene, which provides a summary of the information available at the NCBI for BRCA1. Scroll through the webpage and explore some of the information available. Scroll down the webpage to the section titled “Gene Ontology.” There is a table title ...
Evolution of postzygotic reproductive isolation in galliform birds
... to be increased in the backcrosses. This study contributes to the growing evidence suggesting that the patterns of evolution of postzygotic isolation and the process of speciation are shared among avian groups (and animals in general). In particular, our results support the notion of F2 hybrid invia ...
... to be increased in the backcrosses. This study contributes to the growing evidence suggesting that the patterns of evolution of postzygotic isolation and the process of speciation are shared among avian groups (and animals in general). In particular, our results support the notion of F2 hybrid invia ...
Genes for two multicopper proteins required for Fe(III) oxide
... multicopper proteins found in other micro-organisms because it contains sequences indicative of an Fe(III)binding motif and a fibronectin type III domain. To determine whether ompB expression patterns are diagnostic of physiological responses associated with different metabolic states of Geobacter s ...
... multicopper proteins found in other micro-organisms because it contains sequences indicative of an Fe(III)binding motif and a fibronectin type III domain. To determine whether ompB expression patterns are diagnostic of physiological responses associated with different metabolic states of Geobacter s ...
T -I O -D
... The Winogradsky column creates conditions that expand the volume of natural processes, allowing a clear view of naturally-occurring phenomena. Soil samples are collected from wetland habitats, amended with simple inorganic and organic materials, then exposed to light as an external energy source. Th ...
... The Winogradsky column creates conditions that expand the volume of natural processes, allowing a clear view of naturally-occurring phenomena. Soil samples are collected from wetland habitats, amended with simple inorganic and organic materials, then exposed to light as an external energy source. Th ...
Exploring Host
... the same gene families found in bilaterians and retain many genes that were lost from Drosophila and Caenorhabditis elegans. Because Hydra-associated microbial communities are so complex, fulfilling this analytic challenge will not be easy. Indeed, such communities are distinct from those in the ...
... the same gene families found in bilaterians and retain many genes that were lost from Drosophila and Caenorhabditis elegans. Because Hydra-associated microbial communities are so complex, fulfilling this analytic challenge will not be easy. Indeed, such communities are distinct from those in the ...
BMC Microbiology
... Tyr-specific protein kinases (STYKs) and protein phosphatases, which, as we now know, were encoded in the H. influenzae, M. genitalium, and M. jannaschii genomes [see Additional file 1], see [19,20]. Further, cross-genome comparisons revealed entirely new classes of signaling molecules with GGDEF an ...
... Tyr-specific protein kinases (STYKs) and protein phosphatases, which, as we now know, were encoded in the H. influenzae, M. genitalium, and M. jannaschii genomes [see Additional file 1], see [19,20]. Further, cross-genome comparisons revealed entirely new classes of signaling molecules with GGDEF an ...
Alu - Environmental
... • There is no need for a control because all samples will produce a band therefore it will tell us if the DNA was good without a need for an additional control ...
... • There is no need for a control because all samples will produce a band therefore it will tell us if the DNA was good without a need for an additional control ...
The Gene Gateway Workbook
... The Gene Gateway Workbook is a collection of activities with screenshots and step-by-step instructions designed to introduce new users to genetic-disorder and bioinformatics resources freely available on the Web. It should take about 3 hours to complete all five activities. The workbook activities w ...
... The Gene Gateway Workbook is a collection of activities with screenshots and step-by-step instructions designed to introduce new users to genetic-disorder and bioinformatics resources freely available on the Web. It should take about 3 hours to complete all five activities. The workbook activities w ...
Small Nucleolar RNA
... • Loss of either the paternal or maternal copy of this region leads to two neurological disorders: Prader-Willi or Angelman ...
... • Loss of either the paternal or maternal copy of this region leads to two neurological disorders: Prader-Willi or Angelman ...
How to report IG sequence data in clinical Richard Rosenquist Uppsala, Sweden
... Example laboratory report for IGHV mutation status Determination of immunoglobulin heavy variable (IGHV) gene mutational status in chronic lymphocytic leukemia Patient name: _________________________ ...
... Example laboratory report for IGHV mutation status Determination of immunoglobulin heavy variable (IGHV) gene mutational status in chronic lymphocytic leukemia Patient name: _________________________ ...
Supplementary Methods 1. Generation and post
... Normalized copy number estimates (log2 ratios) were segmented using the Circular Binary Segmentation (CBS) algorithm, followed by median centering the segment values in each sample around 0. These data were obtained from the Level 3 segmented copy number data files that are available for public down ...
... Normalized copy number estimates (log2 ratios) were segmented using the Circular Binary Segmentation (CBS) algorithm, followed by median centering the segment values in each sample around 0. These data were obtained from the Level 3 segmented copy number data files that are available for public down ...
JunctionSeq Package User Manual
... Under the default parameterization, JunctionSeq also builds upon and expands the basic design put forth by DEXSeq, providing (among other things) the ability to test for both differential exon usage and differential splice junction usage. These two types of analyses are complementary: Exons represen ...
... Under the default parameterization, JunctionSeq also builds upon and expands the basic design put forth by DEXSeq, providing (among other things) the ability to test for both differential exon usage and differential splice junction usage. These two types of analyses are complementary: Exons represen ...
Molecular survey and characterization of a novel Anaplasma
... These pathogens exhibit a high degree of biological diversity, broad geographical distribution, and represent a serious threat to veterinary and public health worldwide. Results: A novel Anaplasma species was identified in Haemaphysalis qinghaiensis (Ixodidae) in northwestern China and was molecular ...
... These pathogens exhibit a high degree of biological diversity, broad geographical distribution, and represent a serious threat to veterinary and public health worldwide. Results: A novel Anaplasma species was identified in Haemaphysalis qinghaiensis (Ixodidae) in northwestern China and was molecular ...
E.coli
... Can we use this signal to deduce some more biological information ? We determined the most important metabolic networks in a (translationally biased) organism Can we determine genes belonging to minimal gene sets ? ...
... Can we use this signal to deduce some more biological information ? We determined the most important metabolic networks in a (translationally biased) organism Can we determine genes belonging to minimal gene sets ? ...
Cluster Analysis for Gene Expression Data
... Feature selection is the process of identifying the most effective subset of the original features to use in clustering. Sometimes, one or more transformations may be applied to the original features to produce new salient features. Clustering process This step refers to the application of a cluste ...
... Feature selection is the process of identifying the most effective subset of the original features to use in clustering. Sometimes, one or more transformations may be applied to the original features to produce new salient features. Clustering process This step refers to the application of a cluste ...
Metagenomics

Metagenomics is the study of genetic material recovered directly from environmental samples. The broad field may also be referred to as environmental genomics, ecogenomics or community genomics. While traditional microbiology and microbial genome sequencing and genomics rely upon cultivated clonal cultures, early environmental gene sequencing cloned specific genes (often the 16S rRNA gene) to produce a profile of diversity in a natural sample. Such work revealed that the vast majority of microbial biodiversity had been missed by cultivation-based methods. Recent studies use either ""shotgun"" or PCR directed sequencing to get largely unbiased samples of all genes from all the members of the sampled communities. Because of its ability to reveal the previously hidden diversity of microscopic life, metagenomics offers a powerful lens for viewing the microbial world that has the potential to revolutionize understanding of the entire living world. As the price of DNA sequencing continues to fall, metagenomics now allows microbial ecology to be investigated at a much greater scale and detail than before.