New Reaction Chemistries
... Orthogonality Cost, etc Safety BOC/CBZ/Carbamate Cleavage Redesign protecting groups – enzyme cleavable (penicillin acylase) Aqueous versus organic (recycle) Addition to imines (oxidation) Alternative to Coenzyme A C-C bond formation ...
... Orthogonality Cost, etc Safety BOC/CBZ/Carbamate Cleavage Redesign protecting groups – enzyme cleavable (penicillin acylase) Aqueous versus organic (recycle) Addition to imines (oxidation) Alternative to Coenzyme A C-C bond formation ...
The Central Dogma Dry Lab
... The Central Dogma Dry Lab Following is the base sequence of a gene on one strand of a DNA molecule (the SENSE STRAND): A A T G C C A G T G G T T C G C A C 1. What is the sequence of the complementary DNA strand (i.e. the NONSENSE STRAND)? 2. What is the sequence of the mRNA transcribed from this gen ...
... The Central Dogma Dry Lab Following is the base sequence of a gene on one strand of a DNA molecule (the SENSE STRAND): A A T G C C A G T G G T T C G C A C 1. What is the sequence of the complementary DNA strand (i.e. the NONSENSE STRAND)? 2. What is the sequence of the mRNA transcribed from this gen ...
Unit 3 Review Sheet – Biochemistry
... What are the characteristics of water that make it important to life? Polar, high heat capacity, resists temperature change, abililty to bond and attract other molecules (cohesion and adhesion), ice is less dense than liquid water, universal solvent, most abundant compound in living things What does ...
... What are the characteristics of water that make it important to life? Polar, high heat capacity, resists temperature change, abililty to bond and attract other molecules (cohesion and adhesion), ice is less dense than liquid water, universal solvent, most abundant compound in living things What does ...
Slide ()
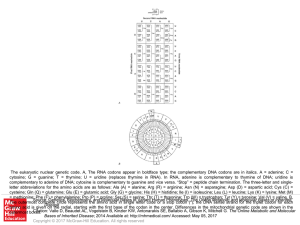
... cytosine; G = guanine; T = thymine; U = uridine (replaces thymine in RNA). In RNA, adenine is complementary to thymine of DNA; uridine is complementary to adenine of DNA; cytosine is complementary to guanine and vice versa. “Stop” = peptide chain termination. The three-letter and singleletter abbrev ...
... cytosine; G = guanine; T = thymine; U = uridine (replaces thymine in RNA). In RNA, adenine is complementary to thymine of DNA; uridine is complementary to adenine of DNA; cytosine is complementary to guanine and vice versa. “Stop” = peptide chain termination. The three-letter and singleletter abbrev ...
Enzymes are specific? - The BioUpdate Foundation
... challenged and there is a danger that it becomes a self perpetuating myth. One of the things we are taught, and which I would like to challenge is the idea that enzymes are specific. I think this idea really is a self perpetuating myth. A biological activity is observed, an enzyme (a biological cata ...
... challenged and there is a danger that it becomes a self perpetuating myth. One of the things we are taught, and which I would like to challenge is the idea that enzymes are specific. I think this idea really is a self perpetuating myth. A biological activity is observed, an enzyme (a biological cata ...
Slide 1
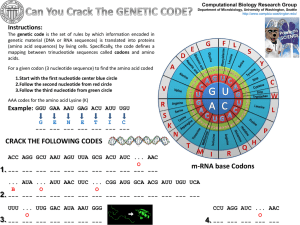
... The genetic code is the set of rules by which information encoded in genetic material (DNA or RNA sequences) is translated into proteins (amino acid sequences) by living cells. Specifically, the code defines a mapping between trinucleotide sequences called codons and amino acids. For a given codon ( ...
... The genetic code is the set of rules by which information encoded in genetic material (DNA or RNA sequences) is translated into proteins (amino acid sequences) by living cells. Specifically, the code defines a mapping between trinucleotide sequences called codons and amino acids. For a given codon ( ...
LOYOLA COLLEGE (AUTONOMOUS), CHENNAI
... (b) Discuss the substrate specificity of enzymes and the different ways in which substrate can be affected when at the active site. 22(a) Describe zymogens with suitable examples. OR (b) Write notes on: (i) Catalytic mechanism of tryptophan synthase (ii) Substrate binding by chymotrypsin and trypsin ...
... (b) Discuss the substrate specificity of enzymes and the different ways in which substrate can be affected when at the active site. 22(a) Describe zymogens with suitable examples. OR (b) Write notes on: (i) Catalytic mechanism of tryptophan synthase (ii) Substrate binding by chymotrypsin and trypsin ...
ภาพนิ่ง 1
... transferase enzymes (e.g. proteases, amidases, esterases, acylases, lipases and β-lactamases). A common method for generating a nucleophilic residue for covalent catalysis is by using an Acid-Base-Nucleophile triad. -Wikipedia- ...
... transferase enzymes (e.g. proteases, amidases, esterases, acylases, lipases and β-lactamases). A common method for generating a nucleophilic residue for covalent catalysis is by using an Acid-Base-Nucleophile triad. -Wikipedia- ...
Biochemistry I, Spring Term 2001 - Second Exam:
... 1. Km and KD are similar in that: a) they refer to the concentration of ligand or substrate in a biochemical process. b) they both relate to ligand binding measurements, Km=1/KD c) they both reflect half-way points in a biochemical process. d) answers a and c. 2. In both hemoglobin and myoglobin the ...
... 1. Km and KD are similar in that: a) they refer to the concentration of ligand or substrate in a biochemical process. b) they both relate to ligand binding measurements, Km=1/KD c) they both reflect half-way points in a biochemical process. d) answers a and c. 2. In both hemoglobin and myoglobin the ...
Structural basis for the functional differences between ASCT1 and
... Structural basis for the functional differences between ASCT1 and EAATs A.J. Scopelliti, R. Ryan and R. Vandenberg, Department. of Pharmacology, Blackburn Building D06, University of Sydney, NSW 2006, Australia. The alanine, serine and cysteine transporters (ASCT1 and 2) are electroneutral exchanger ...
... Structural basis for the functional differences between ASCT1 and EAATs A.J. Scopelliti, R. Ryan and R. Vandenberg, Department. of Pharmacology, Blackburn Building D06, University of Sydney, NSW 2006, Australia. The alanine, serine and cysteine transporters (ASCT1 and 2) are electroneutral exchanger ...
Ch. 8 Enzymes as catalysts Glucokinase is typical enzyme:
... • Lyases diverse cleave C-C, C-O, C-N • Isomerases rearrange, create isomers of starting • Ligases synthesize C-C, C-S, C-O and C-N bonds; Fig. 8.18 ...
... • Lyases diverse cleave C-C, C-O, C-N • Isomerases rearrange, create isomers of starting • Ligases synthesize C-C, C-S, C-O and C-N bonds; Fig. 8.18 ...
The Cell, 5e
... • Lyases diverse cleave C-C, C-O, C-N • Isomerases rearrange, create isomers of starting • Ligases synthesize C-C, C-S, C-O and C-N bonds; ...
... • Lyases diverse cleave C-C, C-O, C-N • Isomerases rearrange, create isomers of starting • Ligases synthesize C-C, C-S, C-O and C-N bonds; ...
PBHS AP Biology
... If the salt concentration is very low, the enzyme will denature and form an inactive precipitate If the salt concentration is very high, new interactions will occur and again an inactive precipitate is formed Intermediate salt concentrations such as human blood (0.9%) is the optimum for many e ...
... If the salt concentration is very low, the enzyme will denature and form an inactive precipitate If the salt concentration is very high, new interactions will occur and again an inactive precipitate is formed Intermediate salt concentrations such as human blood (0.9%) is the optimum for many e ...
Biochemistry PowerPoint
... up chemical reactions without being affected by the reactions themselves. Enzyme: a protein that increases the rate of reactions by lowering the activation energy. ...
... up chemical reactions without being affected by the reactions themselves. Enzyme: a protein that increases the rate of reactions by lowering the activation energy. ...
Review on Biochemistry: Protein Chemistry
... 3. Physical property: aromatic amino acids absorb UV light: 280 nm: Trp > Tyr >> Phe 4. Chemical property: Gly: two ionizable group: NH3+, and COO-; pI = ½ (pK1 + pK2) ...
... 3. Physical property: aromatic amino acids absorb UV light: 280 nm: Trp > Tyr >> Phe 4. Chemical property: Gly: two ionizable group: NH3+, and COO-; pI = ½ (pK1 + pK2) ...
Questions
... 2. Based on results described in question 1, investigators used the technique of sitedirected mutagenesis to synthesize five mutant CK proteins in which the Cys278 residue was replaced with either a Gly, Ser, Ala , Asn or Asp residue. The mutants were called C278G, C278S, C278A, C278N and C278D, re ...
... 2. Based on results described in question 1, investigators used the technique of sitedirected mutagenesis to synthesize five mutant CK proteins in which the Cys278 residue was replaced with either a Gly, Ser, Ala , Asn or Asp residue. The mutants were called C278G, C278S, C278A, C278N and C278D, re ...
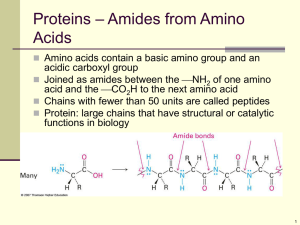
Chapter 26:Biomolecules: Amino Acids, Peptides, and Proteins
... The stereochemical reference for amino acids is the Fischer projection of L-serine Proteins are derived exclusively from L-amino acids ...
... The stereochemical reference for amino acids is the Fischer projection of L-serine Proteins are derived exclusively from L-amino acids ...
Structures and mechanisms
... activities of enzymes are determined by their three-dimensional structure.[20] However, although structure does determine function, predicting a novel enzyme's activity just from its structure is a very difficult problem that has not yet been solved.[21] Most enzymes are much larger than the substra ...
... activities of enzymes are determined by their three-dimensional structure.[20] However, although structure does determine function, predicting a novel enzyme's activity just from its structure is a very difficult problem that has not yet been solved.[21] Most enzymes are much larger than the substra ...
Catalytic triad

A catalytic triad refers to the three amino acid residues that function together at the centre of the active site of some hydrolase and transferase enzymes (e.g. proteases, amidases, esterases, acylases, lipases and β-lactamases). An Acid-Base-Nucleophile triad is a common motif for generating a nucleophilic residue for covalent catalysis. The residues form a charge-relay network to polarise and activate the nucleophile, which attacks the substrate, forming a covalent intermediate which is then hydrolysed to regenerate free enzyme. The nucleophile is most commonly a serine or cysteine amino acid, but occasionally threonine. Because enzymes fold into complex three-dimensional structures, the residues of a catalytic triad can be far from each other along the amino-acid sequence (primary structure), however, they are brought close together in the final fold.As well as divergent evolution of function (and even the triad's nucleophile), catalytic triads show some of the best examples of convergent evolution. Chemical constraints on catalysis have led to the same catalytic solution independently evolving in at least 23 separate superfamilies. Their mechanism of action is consequently one of the best studied in biochemistry.