* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Covert2012_overview
Designer baby wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
RNA interference wikipedia , lookup
Minimal genome wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Microevolution wikipedia , lookup
Gene expression profiling wikipedia , lookup
Nutriepigenomics wikipedia , lookup
RNA silencing wikipedia , lookup
History of RNA biology wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Protein moonlighting wikipedia , lookup
Mir-92 microRNA precursor family wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Point mutation wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Epitranscriptome wikipedia , lookup
Non-coding RNA wikipedia , lookup
Primary transcript wikipedia , lookup
Helitron (biology) wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
RNA-binding protein wikipedia , lookup
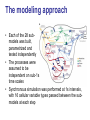
A tremendous modeling feat • Whole-cell, molecular-level computational model of the life-cycle of a procaryote: Mycoplasma genitalium • Integrates 28 different sub-modules, each employing its own modeling technique (ODEs, CBM, Stochastic processes, etc.) Some food for thought Model Simulation Research Engineering Understanding “biological reality” . Understanding “biology as it could be” • What does it mean “to validate” a construct of such complexity? • How could such a tool be used? The modeling approach • Each of the 28 submodels was built, parametrized and tested independently • The processes were assumed to be independent on sub-1s time scales • Synchronous simulation was performed at 1s intervals, with 16 cellular variable types passed between the submodels at each step #genes associated with the process RNA weight, length, composition, etc. Translation progress Status (active/ stalled), RNA species, codon position Fluxes vector Molecule counts for 3 compartments: cytosol, membrane, extracellular space Boolean functions Poisson process ODE system FBA + MOMA Model reconstruction • Chromosome reconstruction: – – – – Genes Transcription units Promoters Protein binding sites • Functional annotation (new annotations for 237 out of 525 genes!) Model reconstruction (cont.) • Structural reconstruction of each gene product: – Protein sequence – Post-transcriptional and post-translational processing and modification of RNA and protein – Signal sequence and localization – DNA footprint of each DNA-binding protein – Chaperones and prosthetic groups required to fold each protein – Subunit composition of each protein and ribonucleoprotein complex – Disulfide bonds of each protein and complex • Curation and complementation of chemical reactions for all processes – The source metabolic model of M. genitalium by Maranas (iPS189 Suthers et al., PLoS CompBio 2009) had 262 reactions and 274 metabolites – The current model has 645 metabolic reactions (out of 1857 reactions overall) and 722 metabolites! ••• Parameter reconciliation • Constraints between parameters in multiple modules were identified • Model parameters were then tuned to – Satisfy these constraints – Deviate minimally from empirical observations Determining initial conditions • Cell proerties right after division were assumed to be statistically identical in consequtive generations • Initial conditions for all cell state variables were iteratively modified, until the postdivision variable distributions converged to a steady state Example: metabolic modeling • Derivation of optimal flux distribution (maximizing biomass) through “modified FBA”: – Network expansion to include the metabolic requirements of the 27 other processes – Internal exchange reactions added to recycle the metabolic byproducts of the other processes – Optimization objective expanded to include the recycling and export of the metabolic byproducts of the other processes – Flux bounds taking into account enzyme copy numbers and catalytic rates • MOMA to then fit the growth rate to the observation (apparently with no distance limitation) • Updating metabolite counts due to transfer reactions between the three compartments Using the simulation • “Wildtype” simulations: – Initializing the cell state from random initial conditions • “Knocked-out strains”: – Set the half-life of the RNA and protein products of the deleted gene to zero – Deleted all RNA and protein products of the deleted gene Movie time… Verification on training data • Simulated 128 wild-type cells in a typical Mycoplasma culture environment, to predict – – • Cellular properties: cell mass; growth rate Molecular properties: count; localization; activity Consistency with observations on: (A) (B) (C) (D) (E) C Doubling time Cellular chemical composition Major cell mass fractions B Gene expression Gene essentiality D E Validation on ‘external’ data sets • The flux through glycolysis is >100-fold more than that through the pentose phosphate and lipid biosynthesis pathways (E) • Predicted metabolite concentrations are within an order of magnitude of those measures in E. coli for all the metabolites in one study and for 70% of them in a more recent study (F) • ‘‘Burst-like’’ protein synthesis due to the local effect of intermittent messenger RNA (mRNA) expression and the global effect of stochastic protein degradation on the availability of free amino acids for translation, which is comparable to recent reports (G) • The mRNA and protein level distributions are consistent with recently reported single-cell measurements (H) Novel predictions (1) Interactions of DNA-binding proteins (A) Predicting chromosomal protein occupancy (averages over 128 wildtype simulations): • • • • (B) (C) Overall average DNA polymerase RNA polymerase Replication Initiator DnaA Temporal dynamics of chromosome exploration Temporal dynamics of gene expression Interactions of DNA-binding proteins (cont.) (D) Spatio-temporal dynamics of DNA and RNA polymerases (E) Collision and displacement frequencies for pairs of DNA-binding proteins (F) Correlation between proteing density and frequency of collision across the chromosome Novel predictions (2): Metabolism as an emergant cell cycle regulator • More cell-to-cell variation in the durations of the replication initiation (64.3%) and replication (38.5%) stages than in cytokinesis (4.4%) or the overall cell cycle (9.4%) • DNA replication proceeds at two distinct rates: – Initially, replication proceeds quickly due to availability of free dNTP in the cell – When the dNTP pool is exhausted the rate limiting factor becomes dNTP synthesis More predictions • Global energy distribution • Molecular pathologies of single-gene disruptions • Model driven discovery: putative alternative NAD electron transfer pathway for • Who knows what more… Great results, but… “Are they real?” ™ ™ Seinfeld: season 4, episode 19 – “The implant” Here are some of the problems • A lot of the model was based on empirical results from other organisms, some of which pretty distant from M. genitalium • The MOMA stage of the metabolic model wasn’t controlled for distance • Due to the complexity of the system, the “External validation” is prone to multiple-hypothesis bias • Validation results don’t necessarily require whole-cell model • No rigorous robustness analysis of the results to perturbations in model parameters (1900 of them…) M. capricolum M. arthritidis