* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Chapter 8
Non-coding DNA wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Epigenetics in learning and memory wikipedia , lookup
Point mutation wikipedia , lookup
Gene expression programming wikipedia , lookup
Human genome wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Gene nomenclature wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Microevolution wikipedia , lookup
Genome evolution wikipedia , lookup
Protein moonlighting wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Designer baby wikipedia , lookup
Nucleic acid tertiary structure wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Gene expression profiling wikipedia , lookup
Helitron (biology) wikipedia , lookup
Short interspersed nuclear elements (SINEs) wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Mir-92 microRNA precursor family wikipedia , lookup
RNA interference wikipedia , lookup
RNA silencing wikipedia , lookup
History of RNA biology wikipedia , lookup
Messenger RNA wikipedia , lookup
Polyadenylation wikipedia , lookup
Non-coding RNA wikipedia , lookup
RNA-binding protein wikipedia , lookup
Alternative splicing wikipedia , lookup
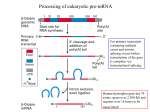
Chap. 8 Post-transcriptional Gene Control Topics • Processing of Eukaryotic Pre-mRNA • Regulation of Pre-mRNA Processing • Cytoplasmic Mechanisms of Post-transcriptional Control Goals • Learn the mechanisms of 5' capping and polyadenylation. • Learn the mechanism of pre-mRNA splicing by the spliceosome complex. • Learn the general functions of splicing repressors and activators in regulation of pre-mRNA splicing. • Learn about mechanisms for translation control via targeted RNA degradation. hnRNP-stained lampbrush chromosome Post-transcriptional Gene Control Post-transcriptional gene control refers to all of the processes that regulate gene expression subsequent to transcription initiation (Fig. 8.1). These processes include regulation of alternative splicing, RNA editing, and RNA degradation. With the exception of alternative splicing, these mechanisms typically are involved in the regulation of only a relatively small fraction of RNAs in a cell. However, they can be highly important for regulation of a given gene. Overview of Pre-mRNA Processing Pre-mRNA processing includes 5' capping, 3’ polyadenylation, and intron splicing (Fig. 8.2). These reactions occur in the nucleus, and begin while the primary transcript is being elongated (cotranscriptional). Mature mRNAs then are transported to the cytoplasm for translation. Eukaryotic pre-mRNA Processing: Capping Bacterial mRNAs are functionally active as transcribed. Eukaryotic pre-mRNAs must be extensively processed to attain their final functional forms. The modification that occurs at the 5' end of the primary transcript is called the 5' cap (m7Gppp) (Fig. 4.14). In this modification, a 7-methylguanylate residue is attached to the first nucleotide of the pre-mRNA by a 5'-5' linkage. The 2'-hydroxyl groups of the ribose residues of the first 2 nucleotides may also be methylated. The 5' cap is important for transport of the mRNA to the cytoplasm, protection against nuclease degradation, and initiation of translation. Mechanism of 5' Capping The synthesis and structure of the 5' cap that is added to most vertebrate mRNAs is illustrated in Fig. 8.3. Caps are added to mRNAs and snRNAs transcribed by RNA Pol II. Capping enzyme removes the phosphate from the 5’ end of the pre-mRNA and adds the 5'-5'-linked guanylate residue to the end of the RNA. Capping enzyme associates with RNA Pol II via its phosphorylated CTD. Other enzymes add the methyl groups to N7 of the 5' guanylate and to 2'hydroxyl groups of the first one or two nucleotides in the primary transcript. (S-Ado-Met: Sadenosylmethionine). Heterogeneous Ribonucleoprotein Particles Pre-mRNAs and other nuclear RNAs are collectively known as heterogeneous nuclear RNA (hnRNA). hnRNA is extensively bound to binding proteins, and complexes between hnRNA and protein are called heterogeneous ribonucleoprotein particles (hnRNP). Binding proteins function by preventing hnRNA from forming tangled 2˚ structures that would otherwise interfere with processing reactions. As illustrated in Fig. 8.5, many RNAbinding proteins contain an RNA recognition motif (RRM) that binds RNA via positively charged amino acids. The cover figure for Chap. 8 shows the extensive hnRNP content of highly transcribed lampbrush chromosomes in newt oocytes. Intro to pre-mRNA Splicing In higher eukaryotes, nearly all genes contain intron sequences that must be spliced out of pre-mRNA to form mature mRNA species. One of the earliest (1977) experiments showing that introns are present in genes is shown in Fig. 8.6. In this experiment, a double-stranded DNA fragment containing most of the adenovirus hexon gene was denatured, hybridized with the hexon mRNA, and then viewed under the electron microscope. As shown in the micrograph and the schematic diagram on the right, DNA loop sequences corresponding to introns removed from the mRNA can be seen looping out from the DNA/RNA hybrid. Splice Site Consensus Sequences Pre-mRNA splice site consensus sequences located at the extreme ends of introns help direct splicing reactions (Fig. 8.7). The identities of these sequences were learned by comparing the sequences of genes to their spliced mRNA products. The GU dinucleotide at the 5' splice site of the intron and the AG dinucleotide at the 3' splice site are highly conserved. Also highly conserved within the intron is a branch point sequence containing the branch-point A residue located ~20-50 nucleotides upstream of the 3' splice site. The remaining central region of the intron (not shown) generally is unimportant for splicing. Mechanism of the Splicing Reaction The splicing reaction occurs via 2 transesterification reactions, for which ∆Gsum ~ 0 (Fig. 8.8). Thus no energy input is required for splicing. In the first reaction, the free 2'-hydroxy group of the branch point A residues attacks and cleaves the phosphodiester linkage at the 5' splice site. In the second reaction, the 3'hydroxy group of the 5' exon attacks and cleaves the phosphodiester linkage at the 3' splice site. The products of the second reaction are the spliced mRNA product and the excised intron, which is called the lariat product. The lariat intron RNA is degraded. Small Nuclear RNAs (snRNAs) and Splicing The splicing reaction requires 5 snRNAs (U1, U2, U4, U5, & U6) that range from about 100-200 nucleotides in length. Each snRNA forms a complex with 6-10 proteins which are called small nuclear ribonucleoprotein particles (snRNPs, pronounced "snurps"). snRNAs bind to pre-mRNA and each other within a larger splicing complex known as the spliceosome (next slide). Interactions between the U1 snRNA and the 5' splice site, and the U2 snRNA and the branch point sequence are crucial in selecting where splicing occurs (Fig. 8.9a). Note that the branch point A residue bulges out of the U2-pre-mRNA duplex. Sm sites indicate where snRNP proteins bind to the snRNAs. Spliceosome Reactions (I) Spliceosomes are large supramolecular complexes consisting of 5 snRNPs and the pre-mRNA. The assembly of the spliceosome and splicing reactions begin with a complex between the pre-mRNA intron, the U1 snRNP bound to the 5' splice site, and the splicing factors SF1 and U2AF bound to the branch point A and pyrimidine tract/3’ AG of the intron, respectively (Fig. 8.11, top ). In Step 1, SF1 departs and the U2 snRNP adds to the complex. In Step 2, the U4/U6/U5 complex adds on forming the fully assembled spliceosome. In Step 3, the U1 and U4 snRNPs depart, and the premRNA is repositioned in the complex for splicing. (Continued on the next slide.) Spliceosome Reactions (II) The transesterification reactions occur in Steps 4 & 5 via the mechanism shown in Fig. 8.11. Following splicing, the remaining components of the complex disassemble. In Step 6, a nuclease known as debranching enzyme cleaves the 2'-5' branch point linkage in the lariat. Degradation of the lariat to individual nucleotides by 3'-to-5' exonucleases then ensues (not shown). It is estimated that ~95% of the polymerized nucleotides within pre-mRNAs ultimately are degraded back to single nucleotides following splicing. RNA Pol II CTD Binds Pre-mRNA Processing Factors Enzymes involved in 5' capping, polyadenylation, and splicing bind to the long phosphorylated CTD of RNA Pol II (Fig. 8.12) while it is transcribing a gene. This ensures that these factors are delivered to the pre-mRNA sites where they are needed. Current research indicates that the binding of these factors to phosphorylated CTD is required to ensure that the enzyme remains processive. Thus, transcription will occur only if these factors are present in sufficient supply. Exon Recognition in Long Pre-mRNAs The average human intron is ~3,500 nucleotides in length, while the average exon is only ~150 nucleotides long. The longest introns are 500 kb in length. As shown in Fig. 8.7, splice site consensus sequences are fairly degenerate, and in long introns, multiple potential 3' acceptor sites occur. Remarkably, exon sequences play an important role in splice site selection in many long introns (Fig. 8.13). Exons contain exonic splicing enhancers (ESEs) that bind SR proteins which recruit the U2 snRNP & U2AF factor to 3' splice sites, and the U1 snRNP to 5' splice sites flanking exons. These assemblies are known as cross-exon recognition complexes. Through this mechanism, the correct splice junctions within a long pre-mRNA are accurately selected. Self-splicing Introns Introns in some protozoan rRNA primary transcripts (group I introns) are self-splicing. Likewise, introns in some protein, rRNA, and tRNA transcripts produced from mitochondrial and chloroplast genes in plants and fungi (group II introns) also carry out selfsplicing reactions. The study of intron self-splicing lead to the discovery of catalytic RNA (ribozymes). Self-splicing introns have strongly conserved secondary and tertiary structures. Because the structure of snRNAs in the spliceosome complex resembles that of group II introns (Fig. 8.14), it is speculated that the spliceosome machinery evolved from group II introns. Early in evolutionary history, when catalytic RNAs may have been much more prevalent, all introns may have been excised by self-splicing. The transfer of splicing reactions to snRNA would have removed constraints on the structure of introns, and thereby facilitated exon shuffling and gene evolution. 3' Cleavage and Polyadenylation of PremRNAs (I) 3' cleavage and polyadenylation of mRNAs are tightly coupled processes that are signaled by 2 sequences near the 3' end of premRNA. These sequences serve as binding sites for 4 nuclear factors (Fig. 8.15). In Step 1, CPSF (cleavage and polyadenylation specificity factor), CStF (cleavage stimulatory factor), and CFI/II (cleavage factors I & II) bind to these sites. Then in Step 2, PAP (poly(A) polymerase) binds to the complex. In Step 3, the premRNA is cleaved just downstream of the AAUAAA poly(A) signal. (Continues on the next slide) 3' Cleavage and Polyadenylation of PremRNAs (II) In Step 4, the CStF, CFI, and CFII factors and the 3' fragment from the pre-mRNA are released. The RNA fragment is rapidly degraded. PAP then begins slow polymerization of the poly(A) tail. In Step 5, PABII (PABPII, poly(A)-binding protein II) adds to the complex and stimulates rapid polymerization of the remainder of the poly(A) tail (Step 6). PABPII also controls the length of the poly(A) tail which typically ranges from 200250 residues. PABPII binds the RNA via a RRM binding sequence. As discussed in Chap. 4, the poly(A) tail functions in translation and mRNA turnover. Intro to Control of Alternative Splicing The most common mechanism by which post-transcriptional gene control is achieved is the regulation of alternative splicing. In humans, ~95% of genes are specified by complex transcription units that produce different protein isoforms due to alternative splicing. Alternative splicing is very common in the nervous system. Alternative splicing is regulated by splicing repressors and activators that control splice site selection. Regulated Splicing in Drosophila Sexual Differentiation (I) One of the best understood systems where alternative splicing is used to regulate gene expression is that used in the control of sexual differentiation in Drosophila embryos. Sexual differentiation is controlled by the sex-lethal (sxl), transformer (tra), and double-sex (dsx) genes (Fig. 8.16). Sxl is a female specific splicing repressor that is not synthesized in males. Sxl not only regulates the splicing of its own primary transcript but also regulates splicing of the premRNA encoding the Tra protein in females. The Sxl and Tra isoforms produced in males are non-functional due to the presence of stop codons in exons 3 and 2 of these respective genes. These exons are skipped in alternative splicing of the sxl and tra transcripts in females. Regulated Splicing in Drosophila Sexual Differentiation (II) Tra protein is a splicing activator. Its expression in females results in the synthesis of the female isoform of Dsx. Its absence in males, results in the synthesis of the male isoform of Dsx. The female form of Dsx is a transcriptional repressor of male differentiation genes. The male form of Dsx is a transcriptional repressor of female differentiation genes. Thus alternative splicing of the sxl gene ultimately determines sex. Mechanism of Action of Tra Protein The Tra splicing activator regulates splice site selection in female embryos by binding to a complex between the Rbp1/Tra2 SR proteins bound to exonic splicing enhancer sequences in the 4th exon of the dsx primary transcript (Fig. 8.17). Binding directs the assembly of the U2 snRNP and the U2AF protein at the 3' end of the intron preceding exon 4. Thus, the 4th exon is spliced into the dsx mRNA in females. This exon is skipped over in splicing of the male dsx transcript. The protein domain encoded by the 4th exon is important in determining the repressor activity of the Dsx TF. Gene Repression by miRNA & siRNA Two post-transcriptional mechanisms for inhibition of gene expression by small single-stranded RNAs were discovered relatively recently in C. elegans. Micro RNAs (miRNAs) inhibit gene expression by blocking the translation of complementary mRNAs. Humans express about 500 miRNAs, and some plants express over 106 miRNAs. Because a single miRNA can bind to more than one target mRNA, it is estimated that about 1/3 of all human genes may be regulated by miRNAs. Short interfering RNAs (siRNAs) inhibit gene expression by specifically targeting a complementary mRNA for degradation. The mechanism of gene silencing by siRNA is known as RNA interference (RNAi) and is an important research tool. RNAi is thought to play a natural role in protection of cells from RNA viruses and retrotransposons. Structures of miRNA & siRNA Both miRNAs and siRNAs are 21-23-nucleotide single-stranded RNAs. miRNAs bind to the 3' UTR regions of complementary mRNAs via imperfect base-pairing (Fig. 8.25a). Thus they often can inhibit translation of more than one mRNA. siRNAs hybridize perfectly without any mismatches to the coding region of their target mRNAs (Fig. 8.25b). Thus they typically regulate only a single mRNA species. Mechanism of Action of mi- and siRNAs miRNAs are produced by the mechanism shown in Fig. 8.26. RNA Pol II transcribes pri-miRNA transcripts that are partially double-helical. The pri-miRNA is processed to a shorter ~ 70 nt pre-miRNA that is then transported to the cytoplasm. The pre-miRNA, which folds into a hairpin structure, is bound by a protein complex containing the enzyme known as Dicer. Dicer cleaves the molecule producing a 21-23-nt double stranded miRNA. Finally, one of the strands is bound by a protein complex known as the RISC complex (RNA-induced silencing complex). The RISC/miRNA complex subsequently binds to the 3’ UTR of a target mRNA leading to its sequestration away from ribosomes. siRNAs are generated in the Dicer reaction from double-helical RNAs introduced into cells or produced from cleavage of viral RNAs. They also are bound by the RISC complex. However, RISC/siRNA complexes bind to the coding region of a target mRNA, and ultimately the target mRNA is cleaved at a site within the perfect siRNA-mRNA duplex (Fig. 8.25b, arrow). RNA Interference (RNAi) In RNA interference, short interfering RNAs (siRNAs, ~21 nts) produced from longer dsRNAs specifically block gene expression by binding to a target mRNA and triggering its degradation. dsRNAs can be transcribed in vitro and injected into an embryo, for example, where processing by the enzyme known as dicer produces the siRNA (Fig. 5.45 a & b). Alternatively, dsRNA can be expressed in vivo in response to some signal. Subsequent processing to siRNA by dicer then triggers mRNA degradation (Fig. 5.45c). RNAi-mediated gene inactivation is commonly applied to silence gene expression in C. elegans, Drosophila, plants, and even mice. The mechanism by which siRNAs cause mRNA degradation is covered in Chap. 8.