* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Nucleic acid recognition from prokaryotes to eukaryotes: Case
Molecular cloning wikipedia , lookup
RNA interference wikipedia , lookup
Protein moonlighting wikipedia , lookup
Gene expression profiling wikipedia , lookup
Community fingerprinting wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
Biochemistry wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Eukaryotic transcription wikipedia , lookup
Alternative splicing wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Nicotinamide adenine dinucleotide wikipedia , lookup
Two-hybrid screening wikipedia , lookup
RNA silencing wikipedia , lookup
List of types of proteins wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Gene regulatory network wikipedia , lookup
Non-coding DNA wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Molecular evolution wikipedia , lookup
Non-coding RNA wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Epitranscriptome wikipedia , lookup
Transcriptional regulation wikipedia , lookup
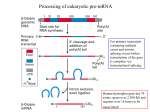
‘Nucleic acid recognition from prokaryotes to eukaryotes: Case studies of a redox-sensing repressor and a pre-mRNA splicing factor’ Clara L. Kielkopf, Assistant Professor University of Rochester School of Medicine Proteins regulate gene expression at multiple stages ranging from transcription through RNA processing and translation. At each stage, regulatory proteins overcome diverse problems of molecular recognition to associate with the target nucleic acid and respond to cellular signals. This seminar describes and contrasts the structural basis for molecular regulation during two distinct cases of gene expression: Structure o f T-Rex/NAD+/DNA. Part (1) NADH/NAD+ redox-sensing by a Rexfamily repressor. Among Gram-positive bacteria, the Rex repressor is a key sensor of oxygen availability that regulates gene expression in response to the intracellular NADH/NAD+ ratio. Here, we present the structure of activated Thermus aquaticus Rex (T-Rex) as a ternary complex with NAD+ and operator DNA at 2.3 Å resolution. Comparison with the previous structure of T-Rex in the NADH-bound state reveals a dramatic conformational rearrangement releases the protein subunits from the DNA in response to NADH. Part (2) Recognition of the 3' pre-mRNA splice site. Almost all human genes contain intervening noncoding introns that must be removed by pre-mRNA splicing. The 3' splice site is marked by consensus sequences, yet variations of these sequences allow specific splice site regulation. Structures of the essential splicing factor U2AF65 bound to a series of splice site variants reveals subtle rearrangements that adapt the protein to different RNA sites. Combined with surface plasmon resonance RNA affinities and small angle X-ray scattering data, we propose a model of adjustable binding registers as a means for universal recognition of diverse 3' splice sites by U2AF65. Altogether, these two examples demonstrate the diversity of conformational changes used by proteins to accomplish gene regulation, from the global rearrangements that dissociate the Rex family repressor from DNA to the subtle rearrangements that adapt U2AF65 to different RNA splice sites.