* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Features of the genetic code
Transcription factor wikipedia , lookup
X-inactivation wikipedia , lookup
Genome evolution wikipedia , lookup
Bottromycin wikipedia , lookup
List of types of proteins wikipedia , lookup
Expanded genetic code wikipedia , lookup
Gene regulatory network wikipedia , lookup
Non-coding DNA wikipedia , lookup
Molecular evolution wikipedia , lookup
Alternative splicing wikipedia , lookup
Point mutation wikipedia , lookup
Biosynthesis wikipedia , lookup
RNA interference wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Messenger RNA wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Polyadenylation wikipedia , lookup
RNA silencing wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
Eukaryotic transcription wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Genetic code wikipedia , lookup
Gene expression wikipedia , lookup
Transcriptional regulation wikipedia , lookup
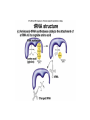
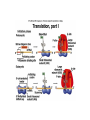
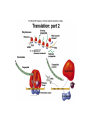
Features of the genetic code: • Triplet codons (total 64 codons) • Nonoverlapping • Three stop or nonsense codons UAA (ocher), UAG (amber) and UGA (opal) • Degenerate • Open reading frame starting at the initiation codon (AUG) • Each codon has 5’ base and a 3’ base e.g. 5’CGU3’ • Mutations that modify the genetic code are of 3 types: frameshift (include deletions and insertions), missense (lead to an amino acid replacement) and nonsense (mutation that generates any of the three stop codons leading a a premature truncation of the polypeptide. TRANSCRIPTION General features: • RNA polymerase (one kind in prokaryote containing five subunits and several kinds in eukaryotes where each transcribes a different type of genes) • Promoter sequences immediately upstream of the gene and contain sequences that have high affinity to bind RNA polymerase • Synthesis of RNA is in the 5’ to 3’ direction. At the transcription bubble, RNA forms a duplex with the template DNA strand where A is complementary to U and C is complementary to G. • Transcription is terminated at the the 3’ end of the gene at sequences know as terminators which direct RNA polymerase to stop synthesis. Processing of RNA transcript in eukaryotes: The immediate RNA transcript is known as primary transcript and is subject to three modifications prior to moving to the cytoplasm: • A capping enzyme adds a G to the first nucleotide in the transcript in the unusual 5’-5’ direction (phosphate to phosphate bond). Then a methyl thransferase adds methyl groups (-CH3) to the G and one or more of the first few bases of the RNA transcript. Capping and methylation is believed to be critical for efficient translation. • Addition of a poly A tail (100-200 As) at the 3’ end of the primary transcript by a poly-A-polymerase. Tailing is believed to stabilize mRNA so it remains for longer time to be translated to many polypeptide molecules before it is degraded and increase the efficiency of the initial steps of translation. • RNA splicing and removal of introns from the primary transcript followed by a very precise joining of the exons. Mechanism of splicing: • Within each intron three sequences are present: splice donors (at the 5’ end of the intron), splice acceptors (at the 3’ end of the intron) and the branch sites (sandwiched between the splice donors and acceptors). • Splicesome is needed to identify and catalyze the sequence of events leading to removal of the intron and rejoining of the two successive exons. The splicesome consists of snRNP (snRNA 100300 nucleotides long + proteins). Each splicesome is composed of four snRNPs together and each snRNP is five snRNA plus about 50 proteins. Some snRNAs base pair with the splice donor and acceptor in the primary transcript and this way can bring them together. • Alternative splicing (as in the gene encoding the antibody heavy chain) and trans-splicing (as in C. elegans) are means of controlling gene expression in eukaryotes.