* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download attachment of amino acids to tRNA
Eukaryotic transcription wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Self-assembling peptide wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
Protein adsorption wikipedia , lookup
List of types of proteins wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Protein (nutrient) wikipedia , lookup
Cell-penetrating peptide wikipedia , lookup
Peptide synthesis wikipedia , lookup
Polyadenylation wikipedia , lookup
Proteolysis wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Gene expression wikipedia , lookup
Protein structure prediction wikipedia , lookup
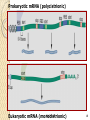
Non-coding RNA wikipedia , lookup
Bottromycin wikipedia , lookup
Biochemistry wikipedia , lookup
Messenger RNA wikipedia , lookup
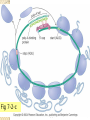
Genetic code wikipedia , lookup
Expanded genetic code wikipedia , lookup
Li Xiaoling Office: QQ: M1623 313320773 E-MAIL: 313320773 @qq.com 2017/5/23 Content Chapter 1 Introduction Chapter 2 The Structures of DNA and RNA Chapter 3 DNA Replication Chapter 4 DNA Mutation and Repair Chapter 5 RNA Transcription Chapter 6 RNA Splicing Chapter 7 Translation Chapter 8 The Genetic code Chapter 9 Regulation in prokaryotes Chapter 10 Regulation in Eukaryotes 2017/5/23 HOW TO LEARN THIS COURSE WELL? To preview and review Problem-base learning Making use of class time effectively Active participation Bi-directional question in class Group discussion Concept map Tutorship To call for reading, thingking and discussing of investigative learning 2017/5/23 To learn effectively EVALUATION (GRADING) SYSTEM in-class and attendance : 10 points Group study and attendance: 20 points Final exam: 70 points Bonus 2017/5/23 Question Ch 5 : Transcription Ch 6 : RNA Splicing Ch 7 : Translation Ch 8 : The Genetic code 4/3/05 Expression of the Genome This part concerned with one of the greatest challenges in understanding the gene-how the gene is expressed. 2017/5/23 6 The revised central dogma Maintenance of the Genome Expression of the Genome RNA processing •Molecular Biology Course CHAPTER 7 Translation What is translation? --it is the story about decoding the genetic information contained in messenger RNA (mRNA) into proteins. 2017/5/23 9 Questions addressed in this chapter What are the main challenges of translation and how do organisms overcome them? What is the organization of nucleotide sequence information in mRNA? What is the structure of tRNAs, and how do aminoacyl tRNA synthetases recognize and attach the correct amino acids to each tRNA? How does the ribosome orchestrate the translation process? 2017/5/23 10 Translation extremely costs In rapid growing bacterial cells, protein synthesis consumes 80% of the cell’s energy 50% of the cell’s dry weight Why? 2017/5/23 11 The main challenges of translation The genetic information in mRNA cannot be recognized by amino acids. The genetic code has to be recognized by an adaptor molecule (translator), and this adaptor has to accurately recruit the corresponding amino acid. 2017/5/23 12 Translation machinery 1. 2. 3. 4. mRNAs (~5% of total cellular RNA) tRNAs (~15%) aminoacyl-tRNA synthetases (氨酰tRNA 合成酶) ribosomes (~100 proteins and 3-4 rRNAs-~80%) 2017/5/23 13 Outline Topics 1-4: Four components of translation machinery. T1-mRNA;T2-tRNA;T3-Attachment of amino acids to tRNA (aminoacyl-tRNA synthetases);T4-The ribosome Topic 5-6:Translation process. T5-initiation;T6-elongation;T7-termination. Topic 8:Translation-dependent regulation of mRNA and protein stability 2017/5/23 14 Topic 1: mRNA Only a portion of each mRNA can be translated. The protein-coding region of the mRNA consists of an ordered series of 3-nt-long units called codons that specify the order of amino acids. 2017/5/23 15 1-1 polypeptide chains are specified by ORF Message RNA The protein coding region of each mRNA is composed of a contiguous, non-overlapping string of codons called an opening reading frame (ORF) . Each ORF begins with a start codon and ends with a stop codon. 2017/5/23 16 The start codon —the first codon of an ORF In bacteria : AUG, GUG, or UUG (5’-3’) In eukaryotic cells: 5’-AUG-3’ Functions: 1.Specifies the first amino acid to be incorporated into the growing polypeptide chain. 2.Defines the reading frame for all subsequent codons. 2017/5/23 17 Prokaryotic mRNA (polycistrionic) 2017/5/23 Eukaryotic mRNA (monocistrionic) 18 Fig 7-1 Three possible reading frames of the E. coli trp leader sequence 2017/5/23 19 Message RNA 1-2 Prokaryotic mRNAs have a ribosome binding site that recruits the translational machinery 1-3 Eukaryotic mRNA are modified at their 5’ and 3’ ends to facilitate translation. 2017/5/23 20 Ribosome binding site (RBS) or SD-sequence in prokaryotic mRNA, complementary with the sequence at the 3’ end of 16S rRNA. Fig 7-2-a structure of prokaryotic mRNA 2017/5/23 21 Once Kozak sequence Fig 7-2-b Eukaryotic mRNA uses a methylated cap to recruit the ribosome. Once bound, the ribosome scans the mRNA in a 5’-3’ direction to find the AUG start codon. Kozak sequence increases the translation efficiency. Poly-A in the 3’ end promotes the efficient recycling of ribosomes. 2017/5/23 22 Fig 7-2-c 2017/5/23 23 Topic 2: tRNA At the heart of protein synthesis is the translation of nucleotide sequence information into amino acids. This work is accomplished by tRNA. 2017/5/23 24 2-1: tRNA are adaptors between codons and amino acids TRANSFER RNA 1. 2. 3. The are many types of tRNA molecules in cell (~40). Each tRNA molecule is attached to a specific amino acids (20) and each recognizes a particular codon, or codons (61), in the mRNA. All tRNAs end with the sequence 5’CCA-3’ at the 3’ end, where the aminoacyl tRNA synthetase adds the amino acid. 2017/5/23 25 Primary structure 1. 2. 3. tRNAs are 75-95 nt in length. There are 15 invariant and 8 semi-invariant residues. The position of invariant and semi-variant nucleosides play a role in either the secondary and tertiary structure. There are many modified bases, which sometimes accounting for 20% of the total bases in one tRNA molecule. Over 50 different types of them have been observed. 2017/5/23 26 4. Pseudouridine (U) is a modified base. These modified bases in tRNA lead to improved tRNA function Fig 7-3 unusual bases 2017/5/23 27 2-2: tRNAs share a common secondary structure that resembles a cloverleaf TRANSFER RNA The cloverleaf structure is a common secondary structural representation of tRNA molecules which shows the base paring of various regions to form four stems (arms) and three loops. 2017/5/23 28 Fig 7-4 the secondary structure 2017/5/23 29 2-3: tRNAs have an L-shaped 3-D structure D loop U loop Fig 7-5 the 3-D structure of tRNA 2017/5/23 30 Formation of the 3-D structure : 9 hydrogen bonds (tertiary hydrogen bonds) mainly involving in the base paring between the invariant bases help the formation of tRNA tertiary structure. 2017/5/23 31 http://www.ncbi.nlm .nih.gov/books/ The cloverleaf structure of a tRNA. The tRNA is drawn in the conventional cloverleaf structure, with the different components labeled. 15 invariant nucleotides (A, C, G, T, U, Y, where Y =pseudouridine) and 8 semi-invariant nucleotides (abbreviations: R, purine; Y, pyrimidine) are indicated. Optional nucleotides not present in all tRNAs are shown as smaller dots. The standard numbering system places position 1 at the 5’ end and position 76 at the 3’ end; not all of the optional nucleotides are included. The invariant and semi-invariant nucleotides are at positions 8, 11, 14, 15, 18, 19, 21, 24, 32, 33, 37, 48, 53, 54, 55, 56, 57, 58, 60, 61, 74, 75 and 76. The nucleotides of the anticodon are at positions 34, 35 and 36. 2017/5/23 32 Base pairing between residues in the D-and Tarms fold the tRNA molecule into an L-shape, with the anticodon loop at one end and the amino acid acceptor site at the other (Fig. 14-5). The base pairing is strengthened by base stacking interactions. 2017/5/23 33 Topic 3: attachment of amino acids to tRNA Amino acids should be attached to tRNA first before adding to polypeptide chain. tRNA molecules to which an amino acid is attached are said to be charged, and tRNAs lacking an amino acid are said to uncharged. 2017/5/23 34 ATTACHMENT OF AMINO ACIDS TO tRNA 3-1 tRNAs are charged by attachment of an amino acid to the 3’ terminal A of the tRNA via a high energy acyl linkage Energy: The energy released when the highenergy bond is broken helps drive the peptide bond formation during protein synthesis. Enzyme: Aminoacyl tRNA synthetase catalyzing the reaction has three binding sites for ATP, amino acid and tRNA. 2017/5/23 35 ATTACHMENT OF AMINO ACIDS TO tRNA 3-2 Aminoacyl tRNA synthetases charge tRNA in two steps (reactions) 1.Adenylylation (腺苷酰化) of amino acids: transfer of AMP to the COO- end of the amino acids. 2. tRNA charging: transfer of the adenylylated amino acids to the 3’ end of tRNA, generating aminoacyl-tRNAs (charged tRNA). 2017/5/23 36 There are two classes of tRNA synthetases. Class I: attach the amino acids to the 2’OH of the tRNA, and is usually monomeric. Class II: attach the amino acids to the 3’OH of the tRNA, and is usually dimeric or tetrameric. 2017/5/23 37 Step 1-Adenylylation of amino acids: the aminoacyl-tRNA synthetase attaches AMP to the-COOH group of the amino acid utilizing ATP to create an aminoacyl (氨酰的) adenylate (腺苷酸) intermediate. As a result, the adenylylated aa binds to the synthetase tightly. [This step is also called activation of amino acids p65] 2017/5/23 38 A class II tRNA synthetase 2017/5/23 39 2017/5/23 40 Step 2- tRNA charging: transfer of the adenylated amino acid to the 3’ end of the appropriate tRNA via the 2’ or 3’OH group, and the AMP is released as a result. 2017/5/23 41 Nature structural and Molecular Biology, 2005, 12:915-922 2017/5/23 42 ATTACHMENT OF AMINO ACIDS TO tRNA 3-3: each aminoacyl tRNA synthetase attaches a single amino acids to one or more cognate/appropriate tRNAs Each of the 20 amino acids is attached to the appropriate tRNA (s) by aminoacyl-tRNA synthetases. Most amino acids are specified by more than one codon, and by more than one tRNA as well. 2017/5/23 43 The same synthetase is responsible for charging all tRNAs for a particular amino acid (one synthetaseone amino acid). Consequently, most organisms have 20 synthetases for 20 different amino acids. 2017/5/23 44 ATTACHMENT OF AMINO ACIDS TO tRNA 3-4 tRNA synthetases recognize unique structure features of cognate tRNAs The recognition has to ensure two levels of accuracy: (1) each tRNA synthetase must recognize the correct set of tRNAs for a particular amino acids; (2) each synthetase must charge all of these isoaccepting tRNAs (即由一种synthetase所识别的不同 tRNAs) 2017/5/23 45 The specificity determinants for accurate recognition are clusters at two distinct sites: the acceptor stem and the anti-codon loop. 2017/5/23 46 Fig 7-8 Fig 7-7 2017/5/23 47 Identity elements (specificity determinants ) in various tRNA molecules 2017/5/23 48 ATTACHMENT OF AMINO ACIDS TO tRNA 3-5 Aminoacyl-tRNA formation is very accurate: selection of the correct amino acid The aminoacyl tRNA synthetases discriminate different amino acids according to different natures of their side-chain groups. 2017/5/23 49 Fig. 7-9 2017/5/23 50 ATTACHMENT OF AMINO ACIDS TO tRNA 3-6 Some aminoacyl tRNA synthetase use an editing pocket to charge tRNAs with high accuracy. 2017/5/23 51 Ile Val Isoleucyl tRNA synthetase as an example: 1. Its editing pocket near the catalytic pocket allows it to proof read the product of the adenylation reaction (step #1). 2. AMP-valine and other mis-bound aa can fit into this editing pocket and get hydrolyzed. But AMP-Ile is too big to fit in the pocket. Thus, the binding pocket serves as a molecular sieve to exclude AMP-valine etc. 2017/5/23 52 Therefore, Ile-tRNA synthetase discriminates against valine twice: the initial binding and adenylylation of the amino acid, and then the editing of the adenylylated amino acid. Each step discriminates by a factor of ~100, and the overall selectivity is about 10,000-fold. 2017/5/23 53 ATTACHMENT OF AMINO ACIDS TO tRNA 3-7 Ribosomes is unable to discriminate between correctly or incorrectly charged tRNAs (是否携带 正确的氨基酸) 1. Ribosome recognize tRNAs but not amino acids (how to prove?). 2. It is responsible to place the charged tRNAs onto mRNA through base pairing of the codon in mRNA and anticodon in tRNA. 2017/5/23 54 Topic 4: the ribosome 1. 2. 3. 4. Ribosome composition Ribosome cycle Peptide bond formation Ribosome structure 2017/5/23 55 4-1 the ribosome is composed of a large and a small subunit RIBOSOMES The large subunit contains the peptidyl transferase center, which is responsible for the formation of peptide bonds. The small subunit interacting with mRNA contains the decoding center, in which charged tRNAs read or “decode” the codon units of the mRNA. 2017/5/23 56 Fig 7-13** Ribosome 2017/5/23 57 RIBOSOMES 4-2: the large and the small subunits undergone association and dissociation during each cycle of translation. 2017/5/23 58 Ribosome cycles: In cells, the small and large ribosome subunits associate with each other and the mRNA, translate it, and then dissociate after each round of translation. This sequence of association and dissociation is called the ribosome cycle. 2017/5/23 59 Fig 7-14 Overview of the events of 2017/5/23 translation/ribosome cycle 60 Polysome/polyribosome: an mRNA bearing multiple ribosomes • Each mRNA can be translated simultaneously by multiple ribosomes Fig 7-15 A polyribosome 2017/5/23 61 2017/5/23 62 RIBOSOMES 4-3 New amino acids are attached to the C-terminus of the growing polypeptide chain. Protein is synthesized in a Nto C- terminal direction 4-4 Peptide bonds are formed by transfer of the growing peptide chain from peptidyltRNA to aminoacyl-tRNA. 2017/5/23 63 Fig 7-16 2017/5/23 64 The structure of the ribosome RIBOSOMES 4-5 Ribosomal RNAs are both structural and catalytic determinants of the ribosomes 4-6 The ribosome has three binding sites for tRNA. 4-7 Channels through the ribosome allow the mRNA and growing polypeptide to enter and/or exit the ribosome. 2017/5/23 65 4-5: Ribosome structure Fig 7-17 two views of the ribosome 2017/5/23 66 4-6 Three binding site for tRNAs A site: to bind the aminoacylated-tRNA P-site: to bind the peptidyl-tRNA E-site: to bind the uncharged tRNA Fig 7-18 2017/5/23 67 Fig 7-19 3-D structure of the ribosome including 3 bound tRNA 2017/5/23 68 4-7 Channel for mRNA entering and exiting are located in the small subunit (see Fig. 14-18) There is a pronounced kink in the mRNA between the two codons at P and A sites. This kink places the vacant A site codon for aminoacyl-tRNA interaction. Fig 7-202017/5/23 69 4-7 Channel for polypeptide chain exiting locates in the large subunit The size of the channel only allow a very limited folding of the newly synthesized polypeptide Fig 7-21 2017/5/23 70 Translation process T5: Initiation of translation T6: Elongation of translation T7: termination of translation Watch the animation on your study CD 2017/5/23 71 Questions 1. 2. Compare the mechanism of translation initiation in prokaryotes and eukaryotes (similarity and difference) How do aminoacyl-tRNA synthetases and the ribosomes contribute to the fidelity of translation, respectively? 2017/5/23 72 Overview of the events of translation Fig 7-14 Initiation Termination Elongation 2017/5/23 73 T5: Initiation of translation Initiation in prokaryotic cells (1-3) Initiation in eukaryotic cells (4-6) 2017/5/23 74 5-1 Prokaryotic mRNAs are initially recruited to the small subunit by base pairing to rRNA. INITIATION OF TRANSLATION Fig 7-23 RBS is also called Shine–Dalgarno sequence 2017/5/23 75 INITIATION OF TRANSLATION 5-2 A specialized tRNA (initiator tRNA) charged with a modified methionine (f-Met) binds directly to the prokaryotic small subunit. Fig 7-24 2017/5/23 76 5-3 Three initiator factors direct the assembly of an initiation complex that contains mRNA and the initiator tRNA. INITIATION OF TRANSLATION 1. Formation of the 30S initiation complex: IFs1-3 + 30S + mRNA + fmettRNA. 2. Formation of the 70S initiation complex: 50S + 30S + mRNA + fmet-tRNA 2017/5/23 77 2017/5/23 78 INITIATION OF TRANSLATION Eukaryotic initiation 5-4 Eukaryotic ribosomes are recruited to the 5’ cap. 5-5 The start codon is found by scanning downstream from the 5’ end of the mRNA. Figs 7-26 and -27 2017/5/23 79 5-6 Translation initiation factors hold eukaryotic mRNAs in circles INITIATION OF TRANSLATION Fig 7-29 Try to explain how the mRNA poly-A tail contributes to the translation efficiency?80 2017/5/23 T6: Translation elongation 1. Aminoacyl-tRNA binding to A site 2. Peptide bond formation 3. Translocation 2017/5/23 81 6-1 Aminoacyl-tRNAs are delivered to the A site by elongation factor EF-Tu ELONGATION OF TRANSLATION 1. EF-Tu-GTP binds to aminoacyl-tRNAs 2. Deliver a tRNA to A site on ribosome 3. When correct codonanticodon occurs, EF-Tu interacts with the factor-binding center on ribosome and hydrolyzes its bound GTP 4. EF-Tu-GDP leaves ribosome 2017/5/23 Fig 7-30 82 6-2 The ribosome uses multiple mechanisms to select against incorrect aminoacyl-tRNAs ELONGATION OF TRANSLATION Additional hydrogen bonds are formed between two adenine residues of the 16S rRNA and the minor groove of the anticodoncodon pair only when they are correctly paired. 2017/5/23 Fig 7-31a 83 Correct base pairing allows EF-Tu interact with the factor binding center on ribosome, which is important for GTP hydrolysis and EF-Tu release. 2017/5/23 Fig 7-31b 84 Only correct base paired aminoacyl-tRNAs remain associated with the ribosome as they rotate into the correct position for peptide bond formation. 2017/5/23 Fig 7-31c 85 6-3 Ribosome is a ribozyme (重点,催化如 何发生?) ELONGATION OF TRANSLATION Fig 7-32 Fig 7-16 2017/5/23 86 ELONGATION OF TRANSLATION 1.The role of L27 protein? 2.The role of A2451 nucleotide residue in the 23 rRNA? 3.The role of the 2’-OH of the A residue at the 3’ of the peptidyl-tRNA (a part of a proton shuttle)? Figure-14-33 2017/5/23 87 ELONGATION OF TRANSLATION 6-4 & 5 Elongation factor EF-G drive translocation of the tRNAs and the mRNA by displacing the tRNA bound to the A site 2017/5/23 88 EF-G mimics a tRNA molecule so as to displace the tRNA bound to the A site Fig 7-35 EF-Tu-GDPNP-Phe-tRNA 2017/5/23 EF-G-GDP 89 ELONGATION OF TRANSLATION 6-6 EF-Tu-GDP and EF-G-GDP must exchange GDP for GTP prior to participating in a new round of elongation. 1. EF-G-GDP: GDP has a lower affinity, and GDP is released after GTP hydrolysis. The free EF-G rapidly binds a new GTP. 2. EF-Tu-GDP requires a GTP exchange factor EF-Ts to displace GDP and recruit GTP to EF-Tu. (Fig. 14-36) 2017/5/23 90 Topic 7:Translation termination 1.Releasing factors act to release the synthesized peptide from the peptidyl tRNA in the ribosome. 2.Ribosome recycling factor, EFTu and IF3 act to recycle the ribosome. 2017/5/23 91 TERMINATION OF TRANSLATION 7-1 & 2 & 3 The action of Class I and II releasing factors 7-1 Class I and Class II releasing factors terminate translation in response to stop codons. 2017/5/23 92 TERMINATION OF TRANSLATION 7-2 Class I releasing factors (bacterial RF1 and RF2, eukaryotic eRF1) recognize stop codons by its peptide anticodon and trigger the release of the peptidyl chain by the conserved GGQ motif.(Figure 14-37). RF1 has a structure resembles tRNA (Figure 14-38), explaining why it can enters A site. 2017/5/23 93 TERMINATION OF TRANSLATION 7-3 Class II releasing factor remove Class I releasing factor from the A site. And this function is controlled by GDP/GTP exchange and GTP hydrolysis (Figure 14-39). 2017/5/23 94 TERMINATION OF TRANSLATION 7-4 Recycling of the ribosome by a combined effort of RRF (ribosome recycling factor), EF-Tu-GTP and IF3. (Figure 14-40) RRF mimics a tRNA and enters A site, and EF-Tu-GTP pushes it into P site. This action pushes the uncharged tRNAs from P-site and E-site. Then mRNA is dissociated from ribosome (on direct interaction), and the small subunit is sequestered by IF3. 2017/5/23 95 Topic 8: translation dependent regulation of mRNA and protein stability Here regulation refers cellular processes that deal with defective mRNA and their translated product. 2017/5/23 96 8-1: The SsrA RNA rescues (拯救) ribosomes that translate broken mRNAs lacking a stop codon (prokaryotes) 1.The ribosomes are trapped or stalled on the broken mRNA lacking a stop codon 2.The stalled ribosomes are rescued by the action of a chimeric RNA molecule that is part tRNA and part mRNA, called tmRNA. 3.SsrA is a 457-nt tmRNA 2017/5/23 97 Fig 7-39 SsrA rescues the stalled ribosomes 2017/5/23 98 8-2: Eukaryotic cells degrade mRNAs that are incomplete or have premature stop codons. Translation is tightly linked to the process of mRNA decay in eukaryotic cells 2017/5/23 99 Nonsense mediated mRNA decay When an mRNA contains a premature stop codon (nonsense codon), the mRNA is rapidly degraded by nonsense mediated mRNA decay. Pre-releasing the ribosome at the nonsense codon prior to reaching the exon-junction complex initiates a talk between the complex and ribosome to remove the 5’ cap from the mRNA 2017/5/23 10 0 1.Translation of a normal eukaryotic mRNA displace all the exon junction complex Fig 7-40a 2017/5/23 10 1 2. Nonsense mediated mRNA decay Fig 7-40b 2017/5/23 10 2 Nonstop mediated decay Non-stop mediated mRNA decay rescues ribosomes that translate mRNAs lacking a stop codon. (1)The lack of a stop codon results in ribosome translation into the poly-A tail to produce poly-Lys at the C-terminus of the polypeptide; the poly-Lys marks the newly synthesized for rapid degradation. 2017/5/23 10 3 (2)The ribosome eventually stalls at the 3’ end of the mRNA, which is bound by the Ski7 protein that triggers the ribosome dissociation and recruits a 3’-5’ exonuclease activity to degrade the “nonstop” mRNA. 2017/5/23 Fig 7-40c 10 4 Key points of the chapter 1. The main challenge of translation 2. The structure and function of four components of the translation machinery. (重点) 3. Translation initiation, elongation and termination (具体过程和翻译因子的作用-注意 起始阶段原核与真核的不同,重点) 4. Translation-dependent regulation of the stability of defective mRNAs and the resulted protein (生物学问题是什么,在原核 和真核分别怎么解决的)2017/5/23 10 5