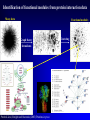
* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Bioinformatik - Brigham Young University
Artificial gene synthesis wikipedia , lookup
Multi-state modeling of biomolecules wikipedia , lookup
Histone acetylation and deacetylation wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Cell-penetrating peptide wikipedia , lookup
Gene regulatory network wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Gene expression wikipedia , lookup
Protein (nutrient) wikipedia , lookup
Protein folding wikipedia , lookup
Homology modeling wikipedia , lookup
Magnesium transporter wikipedia , lookup
Expression vector wikipedia , lookup
Protein moonlighting wikipedia , lookup
Protein structure prediction wikipedia , lookup
List of types of proteins wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Protein domain wikipedia , lookup
Western blot wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
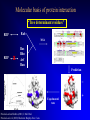
Protein-protein interactions Chapter 12 Stable vs. transient protein-protein interactions Stable complex Transient Interaction Stable complex: homodimeric citrate synthase Transient Signaling Complex Rap1A – cRaf1 Interface 4890 Å2 Hydrophobic interfaces Interface 1310 Å2 “Hydrophilic” interfaces Multi-domain protein Using publicly available interaction data Are there know interaction partners for you pet protein? Check if: 1. There are interactors for your protein in the literature 2. There are databases of interactions where your protein may appear 3. There are homologues of your protein in the protein interaction databases 4. You can predict interactors by other means? 5. This failing, at this point you go back to the bench… Using publicly available interaction data 1. Are there interactors for my protein in the literature ? Problems: •Low coverage •Does not include results from high throughput experiments •Gene names may not be consistent Using publicly available interaction data 2. Are there databases of interactions where my protein may appear? Some DBs: BIND, MINT (General) + organism specific databases (e.g. MIPS/CYGD) Caution! Check: -the experimental methods used to identify the interaction (e.g. high error rate in large scale yeast-two hybrids) -check the method used to incorporate the interaction in the database (e.g. manual curation vs. literature mining using “intelligent” algorithms) Experimental techniques Yeast two-hybrid screens MS analysis of tagged complexes Correlated mRNA expression levels Tagged protein Protein A Protein B Protein C Purified complex with 3 proteins 3 proteins separated by gel electrophoresis 3 proteins identified by mass spectrometry Experimental techniques Yeast two-hybrid screens MS analysis of tagged complexes Correlated mRNA expression levels 90% of genes with conserved co-expression are members of stable complexes Use microarrays to identify co-expression How good is the data? (von Mering et al., Nature 417:399) How good is the data? (von Mering et al., Nature 417:399) ”We estimate that more than half of all current high-throughput interaction data are spurious” Computational prediction of protein interactions Gene fusion events Tryptophan synthetase a b fusion TrpC TrpF 1PII Enright et al (1999) Nature 409:86 Marcotte et al (1999) Science 285: 751 Fused in E.coli Unfused in some other genomes (Synechocystis sp. and Thermotoga maritima.) Computational prediction of protein interactions Phylogenetic profiles Pellegrini et al (1999) PNAS 96: 4285 Computational prediction of protein interactions Pre-computed predictions: where to find them? Identification of functional modules from protein interaction data Messy data Functional modules Graph theory formalisms Pereiral-Leal, Enright and Ouzounis (2003) Proteins in press Custering DIP database • Documents protein-protein interactions from experiment – Y2H, protein microarrays, TAP/MS, PDB • 55,733 interactions between 19,053 proteins from 110 organisms. Organisms # proteins # interactions Fruit fly 7052 20,988 H. pylori 710 1425 Human 916 1407 E. coli 1831 7408 C. elegans 2638 4030 Yeast 4921 18,225 Others 985 401 14 DIP database Duan et al., Mol Cell Proteomics, 2002 • Assess quality – Via proteins: PVM, EPR – Via domains: DPV • Search by BLAST or identifiers / text • URL • Dyrk1a GI 24418935 15 DIP database Duan et al., Mol Cell Proteomics, 2002 • Assess quality – Via proteins: PVM, EPR – Via domains: DPV • Search by BLAST or identifiers / text • Map expression data 16 DIP/LiveDIP Duan et al., Mol Cell Proteomics, 2002 • Records biological state – Post-translational modifications – Conformational changes – Cellular location 17 DIP/Prolinks database Bowers et al., Genome Biol, 2004. • Records functional association using prediction methods: – – – – Gene neighbors Rosetta Stone Phylogenetic profiles Gene clusters 18 Other functional association databases • Phydbac2 (Claverie) • Predictome (DeLisi) • ArrayProspector (Bork) 19 BIND database Alfarano et al., Nucleic Acids Res, 2005 • Records experimental interaction data • 83,517 protein-protein interactions • 204,468 total interactions • Includes small molecules, NAs, complexes • URL 20 BIND database • Displays unique icons of functional classes 21 MPact/MIPS database Guldener et al., Nucleic Acids Res, 2006 • Records yeast protein-protein interactions • Curates interactions: – 4,300 PPI – 1,500 proteins 22 STRING database von Mering et al., Nucleic Acids Res., 2005 • Records experimental and predicted proteinprotein interactions using methods: – – – – Genomic context High-throughput Coexpression Database/literature mining – URL 23 STRING database • Graphical interface for each of the evidence types • Benchmark against Kegg pathways for rankings 24 STRING database • 736,429 proteins in 179 species • Uses COGs and homology to transfer annotation 25 More interaction databases • IntAct (Valencia) – Open source interaction database and analysis – 68,165 interactions from literature or user submissions • MINT (Cesareni) – 71,854 experimental interactions mined from literature by curators – Uses IntAct data model • BioGRID (Tyers) – 116,000 protein and genetic interactions 26 InterDom database Ng et al., Nucleic Acids Res, 2003 • Predicts domain interactions (~30000) from PPIs • Data sources: – – – – Domain fusions PPI from DIP Protein complexes Literature • Scores interactions 27 Definition of CBM • Interacting domain pair – if at least 5 residue-residue contacts between domains (contacts – distance of less than 8 Ǻ) • Structure-structure alignments between all proteins corresponding to a given pair of interacting domains • Clustering of interface similarity, those with >50% equivalently aligned positions are clustered together • Clusters with more than 2 entries define conserved binding mode. 28 DIMA database Pagel et al., Bioinformatics, 2005 • Phylogenetic profiles of Pfam domain pairs • Uses structural info from iPfam • Works well for moderate information content 29 So.. You know your two proteins interact… do you want to know how? Prediction of the molecular basis of protein interactions Molecular basis of protein interaction “Tree determinant residues” Rab REP MSA REP x Ras Rho Arf Ran Prediction _ Experimental tests Pereira-Leal and Seabra (2001) J. Mol. Biol. Pereira-Leal et al (2003) Biochem. Biophys. Res. Com. + Molecular basis of protein interaction “Tree determinant residues” Continued… Sequence Space algorithm AMAS (part of a bigger package) Casari et al (1995) Nat. Struct. Biol 2(2) Molecular basis of protein interaction In silico docking Requires 3D structures of components Conformational changes cannot be considered (rigid body)