* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Russell Group, Protein Evolution
Gene expression wikipedia , lookup
Signal transduction wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Metalloprotein wikipedia , lookup
Paracrine signalling wikipedia , lookup
Expression vector wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Magnesium transporter wikipedia , lookup
Bimolecular fluorescence complementation wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Western blot wikipedia , lookup
Homology modeling wikipedia , lookup
Protein purification wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Interactome wikipedia , lookup
Proteolysis wikipedia , lookup
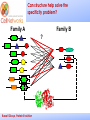
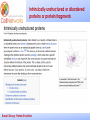
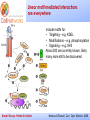
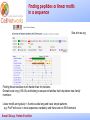
_________ ____ Interactions and Modules: the how and why of molecular interactions Rob Russell Cell Networks University of Heidelberg Russell Group, Protein Evolution _________ ____ Proteins are modular Since the early 1970s it has been observed that protein structures are divided into discrete elements or domains that appear to fold, function and evolve independently. Russell Group, Protein Evolution _________ ____ Domains on a sequence “Low sequence complexity” (Linker regions? Flexible? Junk? Signal peptide (secreted or membrane attached) Transmembrane segment (crosses the membrane) Immunoglobulin domains (bind ligands?) Russell Group, Protein Evolution Tyrosine kinase (phosphorylates Tyr) SMART domain ‘bubblegram’ for human fibroblast growth factor (FGF) receptor 1 (type P11362 into web site: smart.embl.de) _________ ____ Finding domains in a sequence Russell Group, Protein Evolution _________ ____ A library of protein domains for signaling Russell Group, Protein Evolution Pawson & Nash, Science, 2003 _________ ____ Domains assemble to form higher-order structures Russell Group, Protein Evolution Pawson & Nash, Science, 2003 _________ ____ How proteins interact Russell Group, Protein Evolution _________ ____ Modelling interactions by homology homology Protein A X Protein C (e.g.) Two-hybrid interaction Protein D Protein B homology Can we use the C/D structure to predict an interaction between A & B? Russell Group, Protein Evolution Can structure help_________ solve the specificity problem? Family A Family B ? Russell Group, Protein Evolution ____ _________ ____ InterPreTS Interaction Prediction through Tertiary Structure Structure Interface pair potentials Phe ++ Asp Phe Asp Arg -Phe Alignments 1tx4A YFE7_YEAST PIVLRETVAYLQA-------HALTTE ... PLIISSIFSYMDKIYPDLPNDKVR-T ... 1tx4B RHO4_YEAST KLVIVGDGACGKTCLLIVNSKDQF-- ... KIVVVGDGAVGKTCLLISYVQGTFPT ... Score Significance (Do RHO4 & YFE7 interact?) Side-chain to side-chain Side-chain to main-chain Russell Group, Protein Evolution Aloy & Russell, PNAS, 99, 5896, 2002. Aloy & Russell, Bioinformatics. 19, 161, 2003. _________ ____ How proteins interact Russell Group, Protein Evolution _________ ____ Domain peptide interactions Recognition of ligands or targeting signals Post-translational modifications Russell Group, Protein Evolution Linear motifs _________ ____ Peptides interacting with a common domain often show a common pattern or motif usually 3-8 aas. 3BP1_MOUSE/528-537 APTMPPPLPP PTN8_MOUSE/612-629 IPPPLPERTP “instance” SOS1_HUMAN/1149-1157 VPPPVPPRRR NCF1_HUMAN/359-390 SKPQPAVPPRPSA PEXE_YEAST/85-94 MPPTLPHRDW SH3-interacting motif PxxP “motif” “perpetrator” “victim” Russell Group, Protein Evolution Puntervol et al, NAR, 2003; www.elm.org (Eukaryotic Linear Motif DB) _________ ____ Linear motifs versus domains Domains: large globular segments of the proteome that fold into discrete structures and belong in sequence families. Linear motifs: small, non-globular segments that do not adopt a regular structure, and aren’t homologous to each other in the way domains are. Motifs lie in the disordered part of the proteome. Russell Group, Protein Evolution _________ ____ Intrinsically unstructured or disordered proteins or protein fragments Russell Group, Protein Evolution _________ ____ Disorder predictors (IUPred, RONN, DisORPred, etc) Russell Group, Protein Evolution _________ ____ Linear motif mediated interactions are everywhere Include motifs for: • Targeting – e.g. KDEL • Modifications – e.g. phosphorylation • Signaling – e.g. SH3 About 200 are currently known, likely many more still to be discovered Russell Group, Protein Evolution Neduva & Russell, Curr. Opin. Biotech, 2006 Finding peptides _________ or linear motifs in a sequence ____ See: elm.eu.org Finding these modules much harder than for domains. Domains are long (>30 AA) and belong to sequence families that help detect new family members Linear motifs are typically < 8 amino acids long and have simple patterns e.g. PxxP will occur in most sequences randomly and these are not SH3 domains Russell Group, Protein Evolution