* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download protein-protein interactions
Transcriptional regulation wikipedia , lookup
Endomembrane system wikipedia , lookup
Cell-penetrating peptide wikipedia , lookup
Multi-state modeling of biomolecules wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Immunoprecipitation wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Protein (nutrient) wikipedia , lookup
Gene regulatory network wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Gene expression wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Protein folding wikipedia , lookup
Magnesium transporter wikipedia , lookup
Homology modeling wikipedia , lookup
Expression vector wikipedia , lookup
Protein domain wikipedia , lookup
Protein structure prediction wikipedia , lookup
Protein moonlighting wikipedia , lookup
List of types of proteins wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Protein adsorption wikipedia , lookup
Western blot wikipedia , lookup
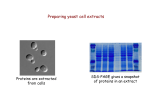
Research Methodology of Biotechnology: Protein-Protein Interactions Yao-Te Huang Aug 19, 2010 The website for downloading lecture notes/lecture materials http://mail.cmu.edu.tw/~ ythuang/teaching.summer .2010.htm Introduction Protein interactions and functions are intimately related. The structure of a protein influences its function by determining the other molecules with which it can interact and the consequences of those interactions. Introduction (contd.) Experimental methods available to detect protein interactions vary in their level of resolution. These observations can be classified into four levels: (a) atomic scale, (b) binary interactions, (c) complex interactions, and (d) cellular scale. Introduction (contd.) Atomic-scale methods: showing the precise structural relationships between interacting atoms and residues The highest resolution methods: e.g., X-ray crystallography and NMR Not yet applied to study protein interactions in a high-throughput manner. Introduction (contd.) Binary-interaction methods: Methods to detect interactions between pairs of proteins Do not reveal the precise chemical nature of the interactions but simply report such interactions take place The major high-throughput technology: the yeast two-hybrid system Introduction (contd.) Complex-interaction methods: Methods to detect interactions between multiple proteins that form complexes. Do not reveal the precise chemical nature of the interactions but simply report that such interactions take place. The major high-throughput technology: systematic affinity purification followed by mass spectrometry Introduction (contd.) Cellular-scale methods: Methods to determine where proteins are localized (e.g., immunofluorescence). It may be possible to determine the function of a protein directly from its localization. COIB (2001), 12:334-339 Principles of proteinprotein interaction analysis These small-scale analysis methods are also useful in proteomics because the large-scale methods tend to produce a significant number of false positives. They include (a) genetic methods, (b) bioinformatic methods, (c) Affinitybased biochemical methods, and (d) Physical methods. Genetic methods Classical genetics can be used to investigate protein interactions by combining different mutations in the same cell or organism and observing the resulting phenotype. Suppressor mutation: A secondary mutation that can correct the phenotype of a primary mutation. Suppressor mutation Synthetic lethal effect Bioinformatic methods (A) The domain fusion method (or Rosetta stone method): The sequence of protein X (a singledomain protein from genome 1) is used as a similarity search query on genome 2. This identifies any singledomain proteins related to protein X and also any multi-domain proteins, which we can define as protein X-Y. As part of the same protein, domain X and Y are likely to be functionally related. The domain fusion method (or Rosetta stone method) The sequence of domain Y can then be used to identify single-domain orthologs in genome 1. Thus, Gene Y, formerly an orphan with no known function, becomes annotated due to its association with Gene X. The two proteins are also likely to interact. The sequence of protein X-Y may also identify further domain fusions, such as protein Y-Z. This links three proteins into a functional group and possibly identifies an interacting complex. The domain fusion method (or Rosetta stone method) Bioinformatic methods (B) The phylogenetic profile: It describes the pattern of presence or absence of a particular protein across a set of organisms whose genomes have been sequenced. If two proteins have the same phylogenetic profile (that is, the same pattern of presence or absence) in all surveyed genomes, it is inferred that the two proteins have a functional link. A protein’s phylogenetic profile is a nearly unique characterization of its pattern of distribution among genomes. Hence any two proteins having identical or similar phylogenetic profiles are likely to be engaged in a common pathway or complex. YPL207W clusters with the ribosomal proteins and can be assigned a function in protein synthesis. When homology is present, the elements are shaped on a gradient from light red (low level of identity) to dark red (high level of identity) Affinity-based biochemical methods (A) Affinity chromatography can be used to trap interacting proteins. If protein X is immobilized on Sepharose beads (e.g., using specific antibodies), then proteins (and other molecules) interacting with protein X can be captured from a cell lysate passed through the column. After washing away unbound proteins, the bound proteins can be eluted, separated by SDS-PAGE and analyzed by mass spectrometry. Affinity chromatography followed by SDS-PAGE & Mass spectrometry Immunoprecipitation The addition of antibodies specific for protein X to a cell lysate will result in the precipitation of the antibodyantigen complex. The technique is usually carried out with polyclonal antisera. The precipitated complexes are separated from the cell lysate by centrifugation, washed and then fractionated by SDS-PAGE, and the bound proteins can be identified by mass spectrometry. Immunoprecipitation GST pulldown The protein X is expressed as a fusion to GST. After mixing the fusion protein with a cell lysate and allowing complexes to form, glutathionecoated beads are added to capture the GST part of the fusion. The beads are recovered by centrifugation, washed and the recovered proteins fractionated and identified by mass spectrometry. GST pulldown Crosslinking Interacting proteins can be identified by crosslinking. A labeled crosslinker is added to protein X in vitro and the cell lysate is added so that interactions can occur. If the crosslink is activated at this stage, interacting proteins become covalently attached to the bait. After purification, the crosslink can be cleaved and the interacting proteins separated by 2D SDS-PAGE. Crosslinking (contd.) Physical methods High-resolution methods: (e.g., X-ray crystallography & NMR) providing data about the relative spacing of atoms of interacting molecules. Low-resolution methods: e.g., electron crystallography and electron tomography. FRET (Fluorescence Resonance Energy Transfer) FRET FRET is the energy transfer that occurs when two fluorophores are close together, and one of fluorophores (the donor) has an emission spectrum that overlaps the excitation spectrum (absorption spectrum) of the other fluorophoe (the acceptor). Basic Theory of FRET: kT(r) = (QD2)(1/Dr6)(9000 *In10)(1/1285NAn4)(∫FD()A() 4d ) = (1/D)(R0/r)6 where R0 is the Förster distance r is the distance between the donor and the acceptor J(), the so-called overlap integral= ∫FD()A() 4d E = 1/(1+(r/R0)6) where E is the efficiency of the energy transfer FRET FRET R0: is the Förster distance FRET: distancedependent R0 is the Förster distance r is the distance between the donor and the acceptor E is the efficiency of the energy transfer Note: when r=R0, E=0.5 FD: the fluorescence intensity of the donor in the absence of the acceptor FDA: the fluorescence intensity of the donor in the presence of the acceptor Library-based methods for the global analysis of binary interactions Standard cDNA expression libraries Phage display method The yeast two-hybrid system Standard cDNA expression libraries Expression libraries are usually screened with labeled antibodies. In place of antibodies, other proteins can be used as probes. For example, labeled calmodulin has been used to screen for calmodulin-binding proteins. Low throughput Does not provide the native conditions for the folding of all proteins, so a significant number of interactions would not be detected. Phage display method (1) M13 (a filamentous phage containing ss-DNA encased in a protein coat): contains five coat proteins, two of which are gVIIIp (gene VIII protein) and gIIIp (gene III protein). Phage display method (2) Phage display method (2): contd. The phage display method The yeast two-hybrid system Transcription factors generally comprise two functionally independent domains, one for DNA binding and one for transcriptional activation. These do not have to be covalently joined together, but can be assembled to form a dimeric protein. This principle is exploited to identify protein interactions. Bait proteins are expressed in one yeast strain as a fusion with a DNA-binding domain and candidate prey proteins are expressed in another strain as fusions with a transactivation domain. When the two strains are mated, functional transcription factors are assembled only if the bait and prey interact. This can be detected by including a reporter gene activated by the hybrid transcription factor. The yeast two-hybrid yeast Limitations of the yeast two-hybrid system First, where independent groups have carried out similar, large-scale studies, the degree of overlap in the reported interactions is very low (1015%). This suggest either that the screens were not comprehensive or that even minor differences in experimental conditions could influence the types of interactions that are detected. Limitations of the yeast two-hybrid system Secondly, a significant number of well-characterized interactions are not detected in the large-scale screens, suggesting there is a high level of false negatives. Thirdly, a significant number of interactions that are detected in large-scale screens appear spurious when investigated in more detail, suggesting there is also high level of false positives. A variant of the yeast two-hybrid system Protein interaction maps Node: proteins or protein complexes are treated as nodes. Edge (or link): interactions between them. Some proteins serve as hubs for very large numbers of interactions. Binary interaction map including 1200 interacting proteins in yeast Trends in Cell biology (2001), 11: 102-106 A simplified version in which yeast proteins have been clustered according to their function Homework Pick up your most favorite method of protein-protein interactions, and describe it in detail. Describe phage display method.