* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download PROTEIN FOLDING
Survey
Document related concepts
Biochemical cascade wikipedia , lookup
Signal transduction wikipedia , lookup
Biochemistry wikipedia , lookup
Gene expression wikipedia , lookup
Paracrine signalling wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Expression vector wikipedia , lookup
Magnesium transporter wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Metalloprotein wikipedia , lookup
Point mutation wikipedia , lookup
Bimolecular fluorescence complementation wikipedia , lookup
Homology modeling wikipedia , lookup
Western blot wikipedia , lookup
Interactome wikipedia , lookup
Protein purification wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Transcript
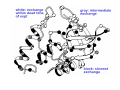
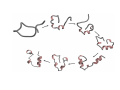
PROTEIN FOLDING Protein folding: the Levinthal Paradox (1968) Assuming 3 possible conformations for each residue An interconversion time of 10-12 sec For a 150-residue protein: 3150 = 1068 possible conformations would be explored after 1056 (1046) years!!!! (more than the age of the universe)...and proteins fold in a few miliseconds! Levinthal’s proposal: There SHOULD be a defined folding pathway through a defined series of intermediates, instead of a fully random process Are there pathways for protein folding? C. Levinthal, J. Chem. Phys. 65, 44 (1968). Unfolded Native K.A. Dill & H.S. Chan, Nature Struct. Biol. 4, 4 (1997). Levinthal Paradox: How do proteins find the native “needle” in the conformational “haystack” in milliseconds when the free energy bias toward the native state is so small? Folding Problem • What structure does a polypeptide fold into? • Unfolded states expose hydrophobic side chains that should be hidden from water, intermolecular interactions of hydrophobic side chains leads to aggregation / precipitation, consequence can be misfolding diseases. • Unfolded states are prone to be digested by proteases. • On the ribosome: How does a 1/2-formed protein chain behave? First report of protein refolding in vitro Anson and Mirsky Alkali-denatured hemoglobin spontaneously recover its biological properties when the pH is returned to neutrality. Anson, M.L. and Mirsky, A. E. (1925) J. Physiol. 60, 50-67. Renaturation of unfolded, denatured ribonuclease C. Anfinsen, 1972 Nobel Laureate The amino acid sequence of a polypeptide chain contains all the information required to fold the chain into its native, 3D structure. Anfinsen, C. B., Haber, E., Sela, M. and White, F. H. (1961) Proc. Natl. Acad. Sci. USA 47, 1309-1314. Sequence determines fold, but.... Protein Folding Related Diseases Natively Unfolded Proteins Why study folding? • What makes a protein stable: kinetic stability or thermodynamic stability? • If we design a new protein, will it fold? • Protein production by overexpression • Protein stabilization by rational mutagenesis • To understand misfolding diseases Flexibility vs. Stability trade-off DePristo, Weinreich & Hartl, Nature Rev Gen. (2005) Thermodynamics of Protein Folding ΔG = ΔH − T Δ S ΔG = RT ln K ΔG fold = ΔGnative − ΔGdenatured ΔGfold is small, about –5 to -20 kcal/mol < kT/10/aa !!!!! therefore the ratio of unfold/fold varies between 0.13 to 0.0003 •Comes from a near balance of opposing large forces •Small forces can therefore play an important role •“Native” state is global minimum of ΔGfold •Folding is reversible (Anfinsen, 1973) The forces that drive folding Reversibility implies ΔGfold is minimized ΔG composed of many contributions • Hydrophobic exclusion (kj/mol) entropic stabilizing • Electrostatics: hydrogen bonding and salt bridges (j/mol) +/- • entropy (degrees of freedom, flexibility) (kj/mol) destabilizing • solvation of polar and charged residues (kj/mol) stabilizing • steric repulsion (kj/mol) destabilizing • van der Waal’s interactions (j/mol) stabilizing Structure and Mechanism in protein science Alan Fersht Energetics of the 3D Structure Experimental Approach: to introduce mutations on a known structure and then monitor stability changes Ala mutations: Smallest possible side chain Gly mutations: to explore flexibility and allowed conformations α-Helix formation • High entropy • Conformational freedom • Reduced steric contacts • Conformational freedom is lost • Intrahelical H bonds entropically favored • Hydrophobic burial Energetics of the 3D Structure β-Sheet formation • Driven by hydrophobic interactions between side chains • β-Branched residues (Thr, Ala) may be favored due to larger degrees of freedom in extended strands • Gly: disfavored (Un)folding experiments • A protein isolated from an organism usually has a well-defined 3D structure (fold). • Changing conditions can unfold the protein: - heat or pressure denaturation - pH denaturation - denaturation by chemicals • Often, but not always, the protein can refold if one takes away the denaturant. • Then, we study the folding transition (kinetics & thermodynamics) Unfolded state • Is the conformation of unfolded proteins random, or is there residual native structure or other structure? We don’t know. • We have a static picture of the F state, don’t know if that is appropriate: Some motions within F must be the motions that can lead to unfolding. • Do different denaturants induce the same unfolded state, or is it U, U’, U’’ … ? What stabilises the fold? • Often a mutation destabilises G only a little • Conclusion: Many interactions add small stability increments. • Isolated helices of proteins don’t fold, or the fold is short-lived, because all interactions added ~kT. • The minimum peptide length that can fold is ~50 amino acids. • But sometimes a few interactions are crucial. • A mutation can have different effects on stability and on folding kinetics! To study protein folding, we need to use different probes To study protein folding, we need to use different probes Es una Serpiente ? Hmmm.. Que rico Purecito!!! Es una Manguera? Es un Arbol? Es una Alfombra? Yeh & Rousseau (2000) Nature Structural Biology 7, 443-445. Table 1. Methods used to investigate protein folding and aggregation Continued Two state model Two-state transition? U (unfolded) ⇔ N (native) Two state-model: • No intermediate accumulation • Cooperative process Intermediates at low concentration may not be detected, giving rise to apparent two-state transitions. U: Unfolded states (ensemble of molecules) May be characterized by IR, CD, NMR, SAXS, hydrodynamics measurements Observation of partially folded species • H-bonded HN in secondary structures of folded proteins exchange slowly with D2O. • Flexible loops get Dlablelled quickly. • If a protein unfolds partly, the HN that disappear fast in D2O belong to the unfolded part. Partially folded species • Some proteins can form partly folded states. • For example, in acid-denatured α lactalbumin, helices B and C are protected from HN exchange. • This means there a partially folded structure can be formed, but does this structure occur during folding? The molten globule • • • • • • • Intermediate found in Compact structure Partially folded May be isolated in some cases Few tertiary contacts Native Secondary structure is retained Fluctuating hydrophobic core There is an energetic barrier to reach the Native state The molten globule Lactalbumin at pH 2 is a molten globule The molten globule The radius of gyration of native α-lactalbumin is 15.7 Å, but the acid molten globule has a radius of 17.2 Å. Molten globule α-lactalbumin retains a globular shape, but is simply ‘swollen’ from the native state (containing 270 bound water molecules). Lactalbumin at pH 2 is a molten globule The molten globule Lactalbumin at pH 2 The molten globule Lactalbumin at pH 2 (molten globule) NH signals exchange faster than in the native state Timescales of experiments for studying protein folding Bieri and Kiefhber, Biol. Chem., 1999 Stopped Flow: how does it work? Excitation Fluo. Cell Mixer A Stopping Syringe B DRIVE Standard stopped flow schematic z Under pneumatic drive activation, the two small volumes of solutions are driven from high performance syringes through a high efficiency mixer. z The resultant mixture passes through a measurement flow cell and into a stopping syringe. z Just prior to stopping, a steady state flow is achieved. z As the solution fills the stopping syringe, the plunger hits a block, causing the flow to be stopped instantaneously. z Using appropriate techniques, the kinetics of the reaction can be measured in the cell. A(t) =ΣAi exp(-kit) + A∞ where A(t) is the amplitude of the change at time t, A∞ is the amplitude at infinite time, Ai is the amplitude at zero time of phase i, and ki is the rate of phase i. For 1-cm window length, 10 m/s flow rate observation time = 0.01/10 = 0.001 s = 1 ms Methods to Study Folding: NMR Folding of Lysozyme Folding models / pathway • What structures form when during folding? • Nucleation / growth: one secondary structure element forms first, then rest attaches to this • Diffusion-collison: all secondary structure form first, then form tertiary structure • Hydrophobic collapse: whole protein become compact first, then secondary structures form • Jigsaw: If a protein folds starting from very different U states, does it fold along the same same path? Figure 19.1 from Fersht Energetics and Kinetics Energetics and Kinetics Energetics and Kinetics The Pathway Solution The bottom of the funnel is thermodynamic minimum Many different kinetic paths reach the bottom Energetics and Kinetics Energetics and Kinetics Protein Folding Funnel Peter Leopold and Jose Onuchic introduced the protein folding funnel approach to the problem. (PNAS 1992) Q=ratio of native contact What do we mean by “folded” protein? The folded state is a collection of conformers that rapidly interconvert since the energy barrier is low at room temp. These energy barriers are small. Structure of the Transition State Solution to the Levinthal Paradox The essence of the solution to the ‘Levinthal Paradox’ that has emerged from the lattice simulations is that an individual molecule needs to sample only a very small number of conformations because the nature of the effective energy surface restricts the search and there are many transition states. Dinner et al., TIBS, 2000, 25, p.331 More Complex Proteins = More Complex Folding: Lysozyme 20% 70% Rapid collapse Qα native contacts in α domain Qβ native contacts in β domain