* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Ultrafast Solvation: Investigating Molecular Forces in Protein Folding November 12, 2010
Survey
Document related concepts
Multi-state modeling of biomolecules wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Phosphorylation wikipedia , lookup
List of types of proteins wikipedia , lookup
Magnesium transporter wikipedia , lookup
Protein moonlighting wikipedia , lookup
Protein design wikipedia , lookup
Rosetta@home wikipedia , lookup
Protein phosphorylation wikipedia , lookup
Circular dichroism wikipedia , lookup
Folding@home wikipedia , lookup
Protein domain wikipedia , lookup
Protein (nutrient) wikipedia , lookup
Protein structure prediction wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Transcript
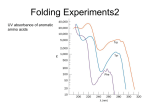
November 12, 2010 Ultrafast Solvation: Investigating Molecular Forces in Protein Folding Christina M. Othon Assistant Professor of Physics Wesleyan University Protein folding is largely driven by non-classical forces such as the hydrophobic effect, van der Waals interactions, and hydrogen bonding. Unlike discrete ionic or covalent bonding, the cooperative behavior of these interactions drives the spontaneous folding and unfolding of large macromolecules. The ability to manipulate these large-scale conformational changes will require a complete understanding of solvent-protein interactions. We investigate solvent-protein interactions by measuring the photoexcitation of the natural amino acid tryptophan using ultrafast fluorescence spectroscopy. This allows us to probe the solvent dynamics at the surface of the molecule. Using this technique we investigated how the variation of solvent properties affects protein conformation. We followed the dynamic hydration changes that took place around an un-folding protein, and found a cooperative unfolding transition involving an intermediate state which is stabilized via long range van der Waals forces. These results provide quantitative results which can help advance protein folding theory and will open new avenues for protein engineering and biotechnology applications.