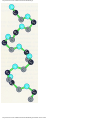
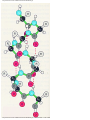
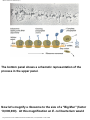
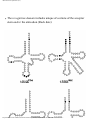
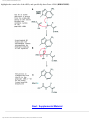
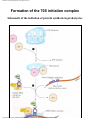
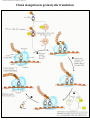
* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Lecture 1 - "Hudel" Luecke
Deoxyribozyme wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Magnesium transporter wikipedia , lookup
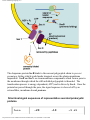
G protein–coupled receptor wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
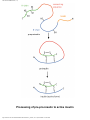
Peptide synthesis wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Interactome wikipedia , lookup
Expression vector wikipedia , lookup
Metalloprotein wikipedia , lookup
Protein purification wikipedia , lookup
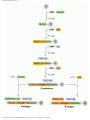
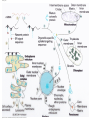
Messenger RNA wikipedia , lookup
Western blot wikipedia , lookup
Point mutation wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Gene expression wikipedia , lookup
Biochemistry wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Transfer RNA wikipedia , lookup
Epitranscriptome wikipedia , lookup
Genetic code wikipedia , lookup
Proteolysis wikipedia , lookup
Lecture 1 Lecture 1 Central Dogma and Deciphering the Genetic Code Hartmut "Hudel" Luecke Department of Molecular Biology and Biochemistry Area of reseach: Structure & Function of Proteins Email: [email protected] http://bass.bio.uci.edu/~hudel/bs99a/lecture20/index.html (1 of 2)5/24/2007 12:49:02 PM Lecture 1 http://bass.bio.uci.edu/~hudel/ bs99a http://bass.bio.uci.edu/~hudel/ bs99b Next: The Central Dogma http://bass.bio.uci.edu/~hudel/bs99a/lecture20/index.html (2 of 2)5/24/2007 12:49:02 PM The Central Dogma (1.1) The Central Dogma DNA 4 nucleotides (A,C,G, T) ==> RNA 4 nucleotides (A,C,G, U) http://bass.bio.uci.edu/~hudel/bs99a/lecture20/lecture1_1.html (1 of 4)5/24/2007 12:49:06 PM ==> Protein 20 amino acids The Central Dogma (1.1) TGGCGAACTGATGTG transcription by UGGCGAACUGAUGUG polymerase phosphodiester bond translation by TrpArgThrAspVal ribosome phosphodiester bond peptide bond Comparing proteins with nucleic acids: Properties proteins have in common with nucleic acid: ● ● Linear heteropolymers with a defined sequence Individual building blocks (called amino acids or simply residues for proteins) are linked together through covalent (chemical) bonds Properties different from nucleic acid: ● ● ● ● More diverse building blocks: 20 amino acids vs. 4 nucleic acids Large variety of functional groups: negatively charged, positively charged, hydrophobic, hydroxyl, sulfhydryl Vastly accelerate a multitude of chemical reactions (also: ribozymes) Assume a wealth of well-defined tertiary structures (shapes): helix bundles, beta sheets, alpha/ beta barrels etc. What do proteins do? ● ● ● ● ● ● Catalyze chemical reactions (enzymes): alcohol dehydrogenase Carry nutrients: hemoglobin is the oxygen carrier in your blood Signaling: peptide hormones bind to protein receptors, transcription factors Molecular recognition: antibodies bind to antigens Play structural roles: finger nails, hair, eye lens Function as motors & pumps: myosin-actin in your muscles, ion pumps http://bass.bio.uci.edu/~hudel/bs99a/lecture20/lecture1_1.html (2 of 4)5/24/2007 12:49:06 PM The Central Dogma (1.1) Proteins are the molecular workhorse of the cell Proteins are of central importance in every cellular process Proteins must be made by the cell with high fidelity Examples of single amino-acid mutations that cause disease: ● ● ● hemoglobin: Glutamic acid (Glu) to valine (Val) mutation at position 6 of the beta chain causes sickle-cell anemia. Fibroblast Growth Factor (FGF) receptor 3: Glycine (Gly) to arginine (Arg) mutation results in achondroplasia, a form of dwarfism for which the gene was discovered here at UCI by Drs. Thompson and Wasmuth. Enzyme uroporphyrinogen III cosynthase: causes congenital porphyria with symptoms such as skin photosensitivity & scarring, mutilating skin deformity, hypertrichosis, hemolytic anemia, red stained teeth. http://bass.bio.uci.edu/~hudel/bs99a/lecture20/lecture1_1.html (3 of 4)5/24/2007 12:49:06 PM The Central Dogma (1.1) Next: Translation Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture20/lecture1_1.html (4 of 4)5/24/2007 12:49:06 PM 3.4 Primary through Quartenary structure Primary Through Quartenary Protein Structure This is a molecule of hexokinase, a metabolic protein found in almost all living organisms. This protein is composed of approximately 6000 atoms and weighs 48 kiloDaltons (kDA). It also has a glucose molecule bound to it, but that is nearly impossible to see. Primary Structure Protein have multiple levels of structure. The most basic is its primary structure. A protein's primary structure is simply the order of its amino acids. Note that this order is always written from amino end to carboxyl end (by convention). An example of a protein primary structure is: http://bass.bio.uci.edu/~hudel/bs99a/lecture20/proteins.html (1 of 6)5/24/2007 12:49:10 PM 3.4 Primary through Quartenary structure 1 31 61 91 121 151 181 211 241 271 301 331 361 391 421 451 A T G S X T X S V A X L I G K X A T S R E X X G C X H R A S P X S R F S F Q V V C K X X A X I S X X L L S A A N X T X X Y R X A 5 D D A A S X D A Q F X X A D I X X D I A X A I A D E A D F Y T X S X V S A F X Y S K X L R S P A L D M M G S D W F N D F L G A 10 V E S A G G X T S V L A S H C D R K S X F X X G V V F S I D V A G T P X G S A A A D C X G H A D A L L I T F K A Q X N E X S L I G X L X P N N G I S G 15 X V I P E V P S F T K L X X I A S L X G V E I A X A A T A A F M I D F I V D P Q N X I X X I V L L X S N A Q S S K C N X V P I W E A Y A I L S T Q X V 20 P P G W X L G N A G M X T D D A X Y R D Y P X M K K N I I X X V A X A N A G X V A K G Y S I L G A K A X X X L K X Y G I L K Y X E X I X T M I V S W A 25 Q A Q V Q E S N X V F P K M G G L N X Y Q K V R S G P Q S S V X S A I A G A X K L R H S Q V G S A K G I G X X P X I A X S S I F G D I X S X H L A X X 30 I A Q A X A S S Q I X X F G M X P X G Q F D F L A X X S X A This is the sequence of hexokinase, yeast hexokinase from the yeast species Saccharomyces cerevisiae to be specific. To find out more about this protein, jump to the Brookhaven Protein Data Bank 3D browser and enter hexokinase in the textbox, or SCOP (Structural Classification of Proteins) and use the PDB reference number 1HKG. Secondary Structure Protein secondary structure refers to certain common repeating structures found in proteins. There are two types of secondary structures: alpha-helix and beta-pleated sheet. An alpha-helix is a tight helix formed out of the polypeptide chain. The polypeptide main chain makes up the central structure, and the side chains extend out and away from the helix. The CO group of one amino acid (n) is hydrogen bonded to the NH group of the amino acid four residues away (n +4). In this way every CO and NH group of the backbone is hydrogen bonded. Here are three models of an alpha-helix. The first shows only the alpha-carbon of each amino acid. The second shows all of the atoms that make up the backbone of the polypeptide. The third shows all of the hydrogen bonds that hold alpha-helices together.The third, and most complete, model is also shown here. http://bass.bio.uci.edu/~hudel/bs99a/lecture20/proteins.html (2 of 6)5/24/2007 12:49:10 PM 3.4 Primary through Quartenary structure A-helices are most commonly made up of hydrophobic amino acids, because hydrogen bonds are generally the strongest attraction possible between such amino acids. a-helices are found in almost all proteins to various extents. For more information read Stryer's Biochemistry, page 27. B-pleated sheets are the other type of secondary structure. They can be either parallel or anti-parallel. Anti-parallel beta-pleated sheets generally look like http://bass.bio.uci.edu/~hudel/bs99a/lecture20/proteins.html (3 of 6)5/24/2007 12:49:10 PM 3.4 Primary through Quartenary structure At the turns they have the structure: where amino acid n hydrogen bonded to amino acid (n +3) in a hairpin turn. There is a special type of molecular model used to highlight protein secondary structure. Follow this link to view an example. This common type of protein model represents segments of beta-pleated sheets as ribbon arrows, and it represents a-helices as helical ribbons. The remainder of the polypeptide chain is referred to as random coil, and is represented as a thin line. Please note that random coil is somewhat of a misnomer; the protein is definitely highly organized, but the random coil regions do not show any easily categorized secondary structure components. http://bass.bio.uci.edu/~hudel/bs99a/lecture20/proteins.html (4 of 6)5/24/2007 12:49:10 PM 3.4 Primary through Quartenary structure Tertiary Structure Tertiary structure is the full 3-dimensional folded structure of the polypeptide chain. There are a number of examples of tertiary in your textbook, and the hexokinase image used as the icon in this module is a complete structure. Quartenary Structure Quartenary structure is only present if there is more than one polypeptide chain. With multiple polypeptide chains, quartenary structure is their interconnections and organization. This is an image of hemoglobin, a protein with four polypeptides-- two alpha-globins, and two beta-globins. The red patches are heme groups (iron complexes bound to the protein, which bind oxygen). http://bass.bio.uci.edu/~hudel/bs99a/lecture20/proteins.html (5 of 6)5/24/2007 12:49:10 PM 3.4 Primary through Quartenary structure http://bass.bio.uci.edu/~hudel/bs99a/lecture20/proteins.html (6 of 6)5/24/2007 12:49:10 PM http://bass.bio.uci.edu/~hudel/bs99a/lecture20/ahelix1.gif http://bass.bio.uci.edu/~hudel/bs99a/lecture20/ahelix1.gif5/24/2007 12:49:12 PM http://bass.bio.uci.edu/~hudel/bs99a/lecture20/ahelix2.gif http://bass.bio.uci.edu/~hudel/bs99a/lecture20/ahelix2.gif5/24/2007 12:49:15 PM http://bass.bio.uci.edu/~hudel/bs99a/lecture20/ahelix3.gif http://bass.bio.uci.edu/~hudel/bs99a/lecture20/ahelix3.gif (1 of 2)5/24/2007 12:49:18 PM http://bass.bio.uci.edu/~hudel/bs99a/lecture20/ahelix3.gif http://bass.bio.uci.edu/~hudel/bs99a/lecture20/ahelix3.gif (2 of 2)5/24/2007 12:49:18 PM 3 Proteins Amino Acids Proteins are very complicated moleucules. With 20 different amino acids that can be arranged in any order to make a polypeptide of up to thousands of amino acids long, their potential for variety is extraordinary. This variety allows proteins to function as exquisitely specific enzymes that compose a cell's metabolism. An E. coli bacterium, one of the most simple biological organisms, has over a 1000 different proteins working at various times to catalyze the necessary reactions to sustain life. 1. Amino Acids Diagram All amino acids have the same general formula: The twenty amino acids found in biological systems are: http://bass.bio.uci.edu/~hudel/bs99a/lecture20/aminoacids.html (1 of 3)5/24/2007 12:49:19 PM 3 Proteins http://bass.bio.uci.edu/~hudel/bs99a/lecture20/aminoacids.html (2 of 3)5/24/2007 12:49:19 PM 3 Proteins All proteins are linear chains composed of these 20 amino acids. http://bass.bio.uci.edu/~hudel/bs99a/lecture20/aminoacids.html (3 of 3)5/24/2007 12:49:19 PM Hammerhead Crystal Structure Hammerhead Ribozyme Crystal Structure An image of the hammerhead ribozyme crystal structure as determined by Pley, Flaherty and McKay ("Three-dimensional structure of a hammerhead ribozyme." Nature (1994), 372: 68-74). The ribozyme is shown bound to an all-DNA substrate inhibitor (in blue), with the 5' end of the substrate (and the 3' end of the ribozyme) on the left of the image. The catalytic core of the ribozyme is shown in red, stem II in purple, and the substrate binding arms of the ribozyme (stems I and III) in green. http://bass.bio.uci.edu/~hudel/bs99a/lecture20/ribozyme.html (1 of 2)5/24/2007 12:49:21 PM Hammerhead Crystal Structure Click here for general information on ribozymes http://bass.bio.uci.edu/~hudel/bs99a/lecture20/ribozyme.html (2 of 2)5/24/2007 12:49:21 PM Translation (1.2) Translation The first step in following the blueprint of DNA to make a protein is transcription which generates an mRNA copy from the DNA template. Translation is the 2nd step: It uses the mRNA template to make the protein polymer. This process is also called protein synthesis. The reasons for having two steps instead of one are: ● ● ● ● Amplification: a single copy gene on DNA can be transcribed into many copies of mRNA Increased levels of control: regulation of transcription as well as translation Ability to separate the mechanisms for DNA replication & transcription from protein synthesis In eukaryotes: Ability to spatially separate replication & transcription (nucleus) from protein synthesis (cytoplasm) http://bass.bio.uci.edu/~hudel/bs99a/lecture20/lecture1_2.html (1 of 2)5/24/2007 12:49:23 PM Translation (1.2) Translation is the process of reading the copy of genetic information on the mRNA (linear sequence of 4 different nucleic acids) and translating it into the proper linear protein sequence of 20 different amino acids. This process is performed in one of the most complex organelles of the cell, the ribosome. In the ribosome the mRNA sequence (information) is read and the corresponding polypeptide (protein) is assembled. The rules for translating the linear nucleic acid sequence (mRNA) into the linear amino acid sequence (protein) are called the Genetic Code. Next: The Genetic Code Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture20/lecture1_2.html (2 of 2)5/24/2007 12:49:23 PM The Genetic Code (1.3) The Genetic Code How to encode a 20-letter alphabet (protein) with a 4letter alphabet (DNA)? 1 nucleotide 4 A, C, G, U 4 amino acids 2 nucleotides 4 x 4 = 16 AU, AG, CA, UU etc. 16 amino acids 3 nucleotides 4 x 4 x 4 = 64 AUG, UGC, CGA etc. 64 amino acids Since two nucleotides are not enough (16), three nucleotides are needed to code for all 20 amino acids Thus Watson & Crick proposed that codon triplets code for individual amino acids. There are several possibilities how triplets might code for amino acids: http://bass.bio.uci.edu/~hudel/bs99a/lecture20/lecture1_3.html (1 of 3)5/24/2007 12:49:25 PM The Genetic Code (1.3) To verify that the Code uses triplets and to determine: ● ● ● overlapping vs. non-overlapping code, punctuated vs. unpunctuated code, the redundancy of the code (64 triplets for 20 amino acids) the following experiments were performed in the early 60s: Next: Experimental Evidence for the Genetic Code Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture20/lecture1_3.html (2 of 3)5/24/2007 12:49:25 PM The Genetic Code (1.3) http://bass.bio.uci.edu/~hudel/bs99a/lecture20/lecture1_3.html (3 of 3)5/24/2007 12:49:25 PM Experimental Evidence (1.4) Experimental Evidence Crick & Brenner (1961) showed the effect of successive deletions of nucleotides in bacteriophage T4 DNA: ATG CTG CTC TGT GCC GCC Original sequence Met Leu Leu Cys Ala Ala ATG CTC TCT GTG CCG CC. 1 nucleotide deleted Met Leu Ser Val Pro Pro ATG CTT CTG TGC CGC C.. 2 nucleotides deleted Met Pro Leu Cys Arg ... ATG CTC TGT GCC GCC ... 3 nucleotides deleted Met Leu Cys Ala Ala ... http://bass.bio.uci.edu/~hudel/bs99a/lecture20/lecture1_4.html (1 of 3)5/24/2007 12:49:26 PM Experimental Evidence (1.4) Deletion of 1 or 2 nucleotides (frame shift mutation) results in non-functional protein Deletion of 3 nucleotides results only in deletion of 1 amino acid (but protein could still be dysfunctional) Insertion of 3 nucleotides results in insertion of 1 amino acid (not shown) Change of 1 nucleotide results in either a sense or silent mutantion or in a missense mutation http://bass.bio.uci.edu/~hudel/bs99a/lecture20/lecture1_4.html (2 of 3)5/24/2007 12:49:26 PM Experimental Evidence (1.4) Nucleotides are read as triplets without overlap or punctuation Next: Deciphering the Genetic Code Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture20/lecture1_4.html (3 of 3)5/24/2007 12:49:26 PM Sequence Mutations Nucleic Acid Sequence Mutations Nonsense Mutation A change in the ribonucleotide base sequence that results in a nonsense (STOP) codon. The protein will be terminated at that point in the message. Missense Mutation This is usually a single substitution mutation and results in one wrong codon and one wrong amino acid. Sense or Silent Mutation This is a single substitution mutation where the change in the DNA base sequence results in a new codon still coding for the same amino acid (Redundacy of the Genetic Code). Frame Shift Mutation When a number of DNA nucleotides not divisible by three is inserted or deleted. This causes a reading frame shift and all of the codons and all of the amino acids after that mutation are usually wrong. Frequently one of the wrong codons turns out to be a nonsense codon and the protein is terminated at that point. Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture20/mutations.html5/24/2007 12:49:27 PM Deciphering the Genetic Code (1.5) Deciphering the Genetic Code Which triplet codon corresponds to which amino acid? Nirenberg (1961) added synthetic homo-polynucleotides to bacterial lysate: poly U UUUUUUUUUUUU Phe-Phe-Phe-Phe poly A AAAAAAAAAAAA Lys-Lys-Lys-Lys poly C CCCCCCCCCCCC Pro-Pro-Pro-Pro Thus the first codon was determined: UUU codes for phenylalanine Alternatively, AAA code for lysine, and CCC for proline. http://bass.bio.uci.edu/~hudel/bs99a/lecture20/lecture1_5.html (1 of 4)5/24/2007 12:49:30 PM Deciphering the Genetic Code (1.5) Khorana (mid 60s) was able to synthesize triplet repeats such as (AAG)n: When AAGAAGAAGAAG was incubated with bacterial lysate containing ribosomes, tRNAs etc., the following results were obtained: Reading frame 1 Reading frame 2 Reading frame 3 -AAG-AAGAAG-AAG- A-AGA-AGAAGA-AG AA-GAA-GAAGAA-G -Lys-LysLys-Lys- -Arg-ArgArg- -Glu-GluGlu- Upon translation, a mixture of homo-polypeptides (poly-lysine, polyarginine and poly-glutamic acid) was obtained, according to the 3 possible reading frames. No hetero-polypeptides were produced, confirming the absence of overlap and punctuation in the Genetic Code. http://bass.bio.uci.edu/~hudel/bs99a/lecture20/lecture1_5.html (2 of 4)5/24/2007 12:49:30 PM Deciphering the Genetic Code (1.5) In 1964 Nirenberg & Leder developed a filter-binding assay: Ribosomes were incubated with one radioactively labeled aminoacyl tRNA* (Trp) and unlabeled (non-Trp) tRNAs. Subsequently, one type of synthetic RNA trinucleotide (AAG or CGA or UGG) was added. When this solution was washed over Millipore filters, only ribosome/tRNA/ mRNA complexes where tRNA* (Trp) AND mRNA (UGG) were complementary remained on the filter. Free, noncomplementary tRNAs and mRNA were washed off. http://bass.bio.uci.edu/~hudel/bs99a/lecture20/lecture1_5.html (3 of 4)5/24/2007 12:49:30 PM Deciphering the Genetic Code (1.5) UGG Trp* tRNA UUU UUC Phe* tRNA In this fashion, the remaining code pairs were determined. Next: The Genetic Code Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture20/lecture1_5.html (4 of 4)5/24/2007 12:49:30 PM Table of the Genetic Code (1.6) Table of the Genetic Code In 1968 Nirenberg & Khorana jointly were awarded the Nobel Prize for the elucidation of the Genetic Code. Noteworthy observations: ● Most codons for a given amino acid differ only in the last (third) base of the triplet (exceptions: Leu, Arg, Ser) http://bass.bio.uci.edu/~hudel/bs99a/lecture20/lecture1_6.html (1 of 2)5/24/2007 12:50:00 PM Table of the Genetic Code (1.6) ● ● One codon (AUG or Met) also signals the START of a polypeptide chain. Three codons (UAA, UAG and UGA) are used to signal the END of a polypeptide chain (STOP codons) For a historical account of the cracking of the Genetic Code click here. Next: Summary Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture20/lecture1_6.html (2 of 2)5/24/2007 12:50:00 PM Summary (1.7) Summary Features of the Genetic Code ● ● ● ● ● ● The Code transfers information from mRNA to proteins with high fidelity It is redundant or degenerate: 61 mRNA triplets code for 20 amino acids Contains START (1) and STOP (3) codons The Genetic Code is nearly universal: correspondence between a nucleotide triplet and an amino acid is identical from viruses to mammals. The rare exceptions are mitochondria and unicellular protozoa. The universality of the Genetic Code is a result of strong evolutionary pressure: a change in a single codon would alter nearly every protein made by an organism. The universality is the basis for recombinant protein technology: mammalian mRNA sequences inserted into bacteria will be correctly expressed (translated). More about this in the last lecture: Expression Systems for Recombinant Proteins. Next lecture: tRNA: Structure & Function http://bass.bio.uci.edu/~hudel/bs99a/lecture20/lecture1_7.html (1 of 3)5/24/2007 12:50:05 PM Summary (1.7) Information Transfer and The Central Dogma DNA -> DNA DNA -> RNA RNA -> Protein Replication Transcription Translation Substrates: dNTPs (A,T,C,G) Substrates: rNTPs (A,U,C,G) Substrates : Amino Acids (20) Chain growth: 5' to 3' Chain growth: 5' to 3' Chain growth: N to C by RNA Polymerase Step 1: tRNA Synthetases (different for each AA) use energy from ATP to couple amino acids to cognate tRNAs. by DNA Polymerase Requires template and primer. Requires only template. Charging specificity determined by 3D structural features unique to each tRNA amino acid pair. Bases added to 3' OH of primer according to WatsonCrick pairing with template. Bases added to 3' OH of growing chain according to Watson-Crick pairing with template. Step 2 by Ribosomes: Initiated at Promoters. Initiated at Start Codon (AUG). In prokaryotes, preceded by ribosome binding site. Two classses of RNAs: Ribosomes provide a platform for binding of tRNA anticodon to individual triplet codons in mRNA according to the Genetic Code. 1. Messenger (mRNA): single stranded, rapid turnover. Amino acids from charged tRNAs are joined to the carboxyl end of the growing chain. Elongation requires GTP hydrolysis. Initiated at replication Origins. In general double stranded and stable. Many mechanisms to assure fidelity during replication (proofreading) and maintenance between replicative rounds (recombination and repair). http://bass.bio.uci.edu/~hudel/bs99a/lecture20/lecture1_7.html (2 of 3)5/24/2007 12:50:05 PM Summary (1.7) 2. Stable RNA (eg. tRNA, ribosomal RNA, snRNAs: folds into compact structures or ribonucleoprotein complexes (RNPs). Processing: Processing: Methylation of bases, ligation Base modification, ligation, of chains, chain cleavage by cleavage, splicing & editing, nucleases. Topological. polyadenylation, 5' capping. Processing: Phosphorylation, acetylation, chain cleavage by proteases, disulfide crosslinking, lipidation, glycosylation etc. Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture20/lecture1_7.html (3 of 3)5/24/2007 12:50:05 PM tRNA: Structure & Function (2.1) Lecture 2 tRNA: Structure & Function Major players in protein synthesis: mRNA, tRNA and the ribosome mRNA Messenger RNA, a copy of DNA blueprint of the gene to be expressed. tRNA Aminoacyl transfer RNA, also called anticodon or adaptor molecule. One or more tRNAs for each amino acid. Supply Ribosome A very large complex of several rRNAs (ribosomal RNA) and many protein molecules. Total molecular weight over 2 million dalton. Factory http://bass.bio.uci.edu/~hudel/bs99a/lecture21/index.html (1 of 4)5/24/2007 12:50:18 PM Information tRNA: Structure & Function (2.1) Protein Polypeptide chain with sequence dictated by the mRNA sequence. Also called the gene product. Product Protein Synthesis Electronmicrograph of a so-called polysome: one mRNA strand (faint horizontal line) with many individual ribosomes attached (dark blobs). The newly synthesized polypeptide chains (proteins) can be seen as irregularly shaped extensions from the ribosomes: http://bass.bio.uci.edu/~hudel/bs99a/lecture21/index.html (2 of 4)5/24/2007 12:50:18 PM tRNA: Structure & Function (2.1) The bottom panel shows a schematic representation of the process in the upper panel. Now let's magnify a ribosome to the size of a "Big Mac" (factor 10,000,000). At this magnification an E. coli bacterium would http://bass.bio.uci.edu/~hudel/bs99a/lecture21/index.html (3 of 4)5/24/2007 12:50:18 PM tRNA: Structure & Function (2.1) be about 10 meters (or 30 feet) in diameter. You would be about 10,000 miles tall: Ribosome 20 nm or 200 Å Big Mac 20 cm 8 inches tRNA 5 nm or 50 Å 5 cm 2 inches mRNA (900 bases) 450 nm or 4,500 Å 450 cm 15 feet Extended (unfolded) protein (300 amino acids) 90 nm or 900 Å 90 cm 3 feet Globular (folded) protein (300 amino acids) 5 nm or 50 Å 5 cm 2 inches 1 nm (nanometer) is 10-9 meters. 1 Å (angstrom) is 10-10 meters or 0.1 nm. Next: tRNA Structure Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture21/index.html (4 of 4)5/24/2007 12:50:18 PM tRNA Structure (2.2) tRNA Structure Primary & secondary structure ● ● ● All tRNAs have similar sequences of 73 to 93 nucleotides 3' end always terminates with the sequence CCA, with the 3' hydroxyl of the ribose of the terminal A being the point of covalent attachment of the amino acid Contain a number (7-15%) of unique/modified bases. These are post-transcriptionally modified after synthesis by RNA polymerase. In particular, adenosine (A) in first or 5' position of the anticodon (corresponding to the third or 3' position of the codon) is always modified to inosine (I) which lacks the amino group on the ❍ ● ● purine ring. Inosine can base-pair with A, U or C and thus accounts for much of the degeneracy of the Genetic Code ("Wobble Theory"). tRNAs have cloverleaf secondary structure due to four base-paired stems The cloverleaf contains three non-base-paired loops: D, anticodon, and TpsiC loop http://bass.bio.uci.edu/~hudel/bs99a/lecture21/lecture2_2.html (1 of 3)5/24/2007 12:50:29 PM tRNA Structure (2.2) Tertiary structure ● ● ● ● ● ● ● The tertiary structure of tRNA is best described as a compact "L" shape. The anticodon is a singlestranded loop at the bottom of the Figure which later basepairs with the triplet codon The amino acid is attached to the terminal A on the upper right. The active sites (anticodon and amino acid) are maximally separated. As in proteins, the tertiary structure is dictated by the primary sequence. The tertiary structure is stabilized by base pairing and base stacking. Two areas (anticodon stem and acceptor stem) form double helix. http://bass.bio.uci.edu/~hudel/bs99a/lecture21/lecture2_2.html (2 of 3)5/24/2007 12:50:29 PM tRNA Structure (2.2) Next: tRNA Function Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture21/lecture2_2.html (3 of 3)5/24/2007 12:50:29 PM tRNA Function: Synthetases (2.3) tRNA Function: Synthetases ● Each tRNA is charged with the proper amino acid via a covalent ester bond at their 3' end by a family of enzymes called aminoacyl-tRNA synthetases. Each enzyme must recognize both the tRNA specific for an amino acid and the corresponding amino acid. This energyconsuming process is ATP-dependent and results in the cleavage of two high-energy phosphate bonds (one in going from ATP to AMP + PP, and one for the cleavage of pyrophosphate into two inorganic phosphates: http://bass.bio.uci.edu/~hudel/bs99a/lecture21/lecture2_3.html (1 of 6)5/24/2007 12:50:39 PM tRNA Function: Synthetases (2.3) ● There are 20 different aminoacyl-tRNA synthetases, one for each amino acid. Despite the fact that they all carry out very similar tasks, they vary greatly in size (40-100 kDalton). http://bass.bio.uci.edu/~hudel/bs99a/lecture21/lecture2_3.html (2 of 6)5/24/2007 12:50:39 PM tRNA Function: Synthetases (2.3) ● Since there are 61 amino acid codons, most tRNA synthetases must be able to recognize more than one type of tRNA (i.e. 6 codons for Arg). These tRNAs are called cognate tRNAs for that particular synthetase. This mapping is achieved through so-called recognition domains on the tRNA. tRNA shown with red backbone and yellow bases. tRNA synthetase shown as space-filling model in blue: http://bass.bio.uci.edu/~hudel/bs99a/lecture21/lecture2_3.html (3 of 6)5/24/2007 12:50:39 PM tRNA Function: Synthetases (2.3) ● The recognition domain includes unique of sections of the acceptor stem and/or the anticodon (black dots): http://bass.bio.uci.edu/~hudel/bs99a/lecture21/lecture2_3.html (4 of 6)5/24/2007 12:50:39 PM tRNA Function: Synthetases (2.3) Accuracy & Proofreading ● ● The accuracy of charging tRNA with the proper amino acid is crucial because once charged, only the tRNA anticodon determines incorporation, not the attached amino acid. The error rate of charging is very low: 1 in 10,000. This is achieved by two means: ❍ ❍ the amino acid specificity pocket in a specific synthetase will only bind amino acids similar in size and charge. the synthetase also has http://bass.bio.uci.edu/~hudel/bs99a/lecture21/lecture2_3.html (5 of 6)5/24/2007 12:50:39 PM tRNA Function: Synthetases (2.3) proofreading capability which, once a wrong aminoacyladenylate complex is formed (1st step), will hydrolyze the complex before it can be covalently attached to the tRNA (2nd step). Next: The Wobble Theory Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture21/lecture2_3.html (6 of 6)5/24/2007 12:50:39 PM The Wobble Theory (2.4) The Wobble Theory Assumption: Each tRNA, defined by its 3-base anticodon, pairs only with its complementary codon on the mRNA. Discrepancy: tRNAAla was found to bind to codons GCA, GCC and GCU. Codon (5'->3') Anticodon (3'->5') GCA CGU http://bass.bio.uci.edu/~hudel/bs99a/lecture21/lecture2_4.html (1 of 4)5/24/2007 12:50:47 PM GCC CGG GCU CGA The Wobble Theory (2.4) http://bass.bio.uci.edu/~hudel/bs99a/lecture21/lecture2_4.html (2 of 4)5/24/2007 12:50:47 PM The Wobble Theory (2.4) The answer: The tRNAAla anticodon is actually CGI which pairs with all three codons. Experimentally determined codon-anticodon pairing rules: ● ● ● The first two positions of the mRNA codon observe Watson-Crick base pairing rules (A-U, C-G) The third position exhibits wobble. Wobble occurs because the conformation of the tRNA anticodon loop permits flexibility at the first base of the anticodon. 5' anticodon base (tRNA) A (not observed) C 3' codon base (mRNA) U (Watson-Crick) G (Watson-Crick) G C or U U A or G http://bass.bio.uci.edu/~hudel/bs99a/lecture21/lecture2_4.html (3 of 4)5/24/2007 12:50:47 PM The Wobble Theory (2.4) I A or C or U Caveat: Since a single tRNA can respond to more than one codon, one tRNA could respond to codons for two different amino acids! This would lead to an ambiguous code. But since the Genetic Code is unambiguous, certain anticodons are disallowed. Next: The Dangers of Wobble Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture21/lecture2_4.html (4 of 4)5/24/2007 12:50:47 PM The Dangers of Wobble (2.5) The Dangers of Wobble Since a single tRNA can respond to more than one codon, one tRNA could in theory respond to codons for two different amino acids: AGC and AGU both code for Ser. Their anticodon is UCG using wobble rules. But anticodon UCA would also pair with AGU (Ser). This anticodon (UCA) would be converted post-transcriptionally to UCI (Hey, that's us!) which would recognize AGC and AGU as intended, but also AGA which codes for Arg -- not good, Example: would result in an ambiguous Genetic Code! One anticodon for serine is UCG: Codon (5'->3') Anticodon (3'->5') AGC UCG AGU UCG Amino acid Ser Ser (wobble) Now, in the following hypothetical example, anticodon UCA (if it existed) would also base-pair with codon AGU using wobble rules: AGU AGU UCA => UCI Ser Ser (wobble) AGC UCI AGU UCI AGA UCI Ser Ser Arg http://bass.bio.uci.edu/~hudel/bs99a/lecture21/lecture2_5.html (1 of 2)5/24/2007 12:50:52 PM (wobble) The Dangers of Wobble (2.5) But anticodon UCA would be converted post-transcriptionally to UCI which is able to bind three codons (wobble rules), including one for Arg! For this reason the UCA/UCI anticodon does not exist! And the codon AGA for Arg is actually covered by the anticodon UCU (wobble)! Next: Supplemental Material Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture21/lecture2_5.html (2 of 2)5/24/2007 12:50:52 PM Supplemental Material (2.6) Supplemental Material This page won't be on the final! Molecular Graphics: This exercise will let you get a better 3-dimensional picture of the interaction of a tRNA with its tRNA synthetase. You will be able to rotate, scale and otherwise manipulate the structure on your screen. First, you'll need to obtain & install a FREE molecular graphics program called RASMOL on your PC: Click here to obtain the MS Windows/NT version or Click here to obtain the Apple Macintosh version Unpack the distribution, you should then be able to run the program RASWIN (PC) or RASMAC (MAC). Click here for detailed help. If you get stuck or need help, please contact me (Hudel). Second, you'll need to get a copy of the atomic coordinates from the Protein Data Bank: PDB entry 1GTR Save this rather large file (0.5 MBytes) with the "Save full entry to disk" option. Save the file as "1GTR.pdb". This file contains the complete atomic coordinates of GLUTAMINYL-tRNA SYNTHETASE COMPLEXED WITH tRNA AND ATP. http://bass.bio.uci.edu/~hudel/bs99a/lecture21/lecture2_6.html (1 of 2)5/24/2007 12:51:02 PM Supplemental Material (2.6) Now, run the program RASWIN/RASMAC and go to the "Open..." option under the "File" Menu. Select the file you just saved (1GTR.pdb). Then select the "Backbone" option under the "Display" menu. Your display should show something like this. For a description of the program, check the "User Manual" under the "Help" Menu. Have fun! Next: Summary Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture21/lecture2_6.html (2 of 2)5/24/2007 12:51:02 PM Summary (2.7) Summary Features of tRNAs ● ● ● ● ● ● ● tRNAs are the adaptor molecules that translate from nucleic acid triplet to amino acid. tRNAs are charged with amino acids by enzymes called aminoacyltRNA synthetases in an energy-dependent 2-step process. There are 20 specific synthetases, one for each amino acid. Since most amino acids have more than one possible codon, there are more than 20 different tRNAs (40 in E. coli). Not all 61 codons have a specific tRNA due to wobble in the 3rd codon position. tRNA synthetases highly specific and have proofreading capabilities, achieving a better than 1:10,000 error rate. The accuracy of charging tRNA with the proper amino acid is crucial because once charged, only the tRNA anticodon determines incorporation into the growing polypeptide chain, not the attached amino acid. Next Lecture: The Ribosome, rRNA and mRNA Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture21/lecture2_7.html5/24/2007 12:51:06 PM The Ribosome, rRNA and mRNA (3.1) Lecture 3 The Ribosome, rRNA and mRNA Major players in protein synthesis: mRNA, tRNA and the ribosome mRNA Messenger RNA, a copy of DNA blueprint of the gene to be expressed. tRNA Aminoacyl transfer RNA, also called anticodon or adaptor molecule. One or more tRNAs for each amino acid. Supply Ribosome A very large complex of several rRNAs (ribosomal RNA) and many protein molecules. Total molecular weight almost 3 million dalton. Factory Protein Polypeptide chain with sequence dictated by the mRNA sequence. Also called the gene product. Product The Ribosome http://bass.bio.uci.edu/~hudel/bs99a/lecture22/index.html (1 of 4)5/24/2007 12:51:15 PM Information The Ribosome, rRNA and mRNA (3.1) Ribosomes can be found either free in the cytosol (cytoplasm) or attached to intracellular membranes. Free ribosomes ● ● ● ● Found in the cytosol. May occur as a single ribosome or in groups known as polyribosomes or polysomes. Occur in greater number than bound ribosomes in cells that retain most of their manufactured protein. Responsible for proteins that go into solution in the cytoplasm or form important cytoplasmic structural or motile elements. http://bass.bio.uci.edu/~hudel/bs99a/lecture22/index.html (2 of 4)5/24/2007 12:51:15 PM The Ribosome, rRNA and mRNA (3.1) Bound ribosomes ● ● ● Found bound to the exterior of the endoplasmic reticulum (ER) constituting the rough ER. Occur in greater number than free ribosomes in cells that secrete their manufactured proteins (e.g., pancreatic cells, producers of digestive enzymes). Responsible for proteins that insert into membranes or are packaged into vesicles for storage in the cytoplasm or export to the cell exterior. Electronmicrograph of ribosomes (black dots) attached to the rough endoplasmic reticulum. ● ● ● ● ● ● About 15,000 ribosomes in a single E. coli cell, comprising ~25% of the dry cell mass. All ribosomes within one cell are identical. All ribosomes are composed of two subunits (called small and large). A substantial fraction of ribosomes are dissociated into free subunits in the cell. Prokaryotic and eukaryotic ribosomes differ in composition. Mitochondrial ribosomes resemble prokaryotic ribosomes. Next: The Composition of Ribosomes http://bass.bio.uci.edu/~hudel/bs99a/lecture22/index.html (3 of 4)5/24/2007 12:51:15 PM The Ribosome, rRNA and mRNA (3.1) Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture22/index.html (4 of 4)5/24/2007 12:51:15 PM The Composition of Ribosomes (3.2) The Composition of Ribosomes Prokaryotic ribosomes ● Sedimentation coefficient: 70S ❍ ❍ small subunit: 30S ■ One rRNA molecule (16S) ■ 21 different proteins, designated S1-S21 large subunit: 50S ■ Two rRNA molecules (5S and 23S) ■ 31 different proteins, designated L1-L31 (L12 is present in four copies) http://bass.bio.uci.edu/~hudel/bs99a/lecture22/lecture3_2.html (1 of 4)5/24/2007 12:51:24 PM The Composition of Ribosomes (3.2) The sedimentation coefficient is measured in Svedberg units (S): the rate of sedimentation of a component in a centrifuge is related both to the molecular weight and the 3-D shape of the component. The rRNA components of the prokaryotic ribosome Type Approximate number of nucleotides Subunit location 16S 1,542 30S 5S 120 50S 23S 2,904 50S RNA function and turnover in the cell Nucleotides % of Synthesis % of Total RNA Stability Thousands 500-6000 40-50 3 T1/2 = 1-3 min 3 (23S, 16S, 5S) 2904, 1542, 120 50 90 Stable ~50 73-93 3 7 Stable Different Kinds mRNA Messenger rRNA Ribosome Type tRNA Function Adapter http://bass.bio.uci.edu/~hudel/bs99a/lecture22/lecture3_2.html (2 of 4)5/24/2007 12:51:24 PM The Composition of Ribosomes (3.2) Separation of ribosomal proteins by 2D gel electrophoresis (a) small (30S) subunit; (b) large (50S) subunit http://bass.bio.uci.edu/~hudel/bs99a/lecture22/lecture3_2.html (3 of 4)5/24/2007 12:51:24 PM The Composition of Ribosomes (3.2) In 1968, Dr. M. Nomura, a professor here at UCI, was the first to show that the 30S subunit could be reassembled from the individual components. He found that the order of addition of components was critical. Eukaryotic ribosomes ● Sedimentation coefficient: 80S ❍ ❍ small subunit: 40S ■ One rRNA molecule (18S) ■ 33 different proteins, designated S1-S33 large subunit: 60S ■ Three rRNA molecules (5S, 5.8S, and 28S) ■ 50 different proteins, designated L1-L50 The rRNA components of the eukaryotic ribosome Type Approximate number of nucleotides Subunit location 18S 1,900 40S 5S 120 60S 5.8S 156 60S 4,700 60S 28S Next: Simplified Overview of Translation Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture22/lecture3_2.html (4 of 4)5/24/2007 12:51:24 PM Simplified Overview of Translation (3.3) Simplified Overview of Translation 1. Formation of the initiation complex 2. Elongation of the polypeptide chain (one repetition of the steps a, b and c for every amino acid incorporated into the protein being synthesized): a: binding of the aminoacyl-tRNA b: peptide bond formation c: translocation 3. Termination Rate of synthesis http://bass.bio.uci.edu/~hudel/bs99a/lecture22/lecture3_3.html (1 of 2)5/24/2007 12:51:30 PM Simplified Overview of Translation (3.3) ● ● Elongation is the rate limiting step in protein synthesis In E. coli at 37 degrees C: ❍ ribosome passes through 15 codons per second ❍ 300 amino acid polypeptide made in 20 seconds ❍ 15,000 ribosomes per cell can make 750 molecules of a 300 amino acid protein per second Next: Structure of the Ribosome Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture22/lecture3_3.html (2 of 2)5/24/2007 12:51:30 PM The Structure of the Ribosome (3.4) The Structure of the Ribosome (Prokaryotic ribosomes) Proposed secondary structure of 16S rRNA http://bass.bio.uci.edu/~hudel/bs99a/lecture22/lecture3_4.html (1 of 6)5/24/2007 12:51:37 PM The Structure of the Ribosome (3.4) ● ● Many regions are self-complementary and capable of forming double helical segments Secondary structure is more highly conserved than primary sequence, i.e. complementary mutations evolve to maintain base paring. 3-Dimensional structure of the 70S ribosome http://bass.bio.uci.edu/~hudel/bs99a/lecture22/lecture3_4.html (2 of 6)5/24/2007 12:51:37 PM The Structure of the Ribosome (3.4) The large subunit has a tunnel about 10 nm long and 2.5 nm in diameter. This tunnel is thought to be the channel that newly assembled polypeptide chains pass through on their way out of the ribosome. The two subunits interact very tightly and form the active site http://bass.bio.uci.edu/~hudel/bs99a/lecture22/lecture3_4.html (3 of 6)5/24/2007 12:51:37 PM The Structure of the Ribosome (3.4) Different views: in yellow the 30S (small) subunit; http://bass.bio.uci.edu/~hudel/bs99a/lecture22/lecture3_4.html (4 of 6)5/24/2007 12:51:37 PM in blue the 50S (large) subunit. The Structure of the Ribosome (3.4) Landmarks of the 30S subunit: h, head; p, platform; ch, channel presumed to be the conduit for mRNA; sp, spur. Landmarks of the 50S subunit: CP, central protuberance; St, L7/L12 stalk; L1, L1 protein; IC, interface canyon; T, tunnel presumed to be the conduit for the nascent polypeptide chain; T1 and T2, lower tunnel http://bass.bio.uci.edu/~hudel/bs99a/lecture22/lecture3_4.html (5 of 6)5/24/2007 12:51:37 PM The Structure of the Ribosome (3.4) segments, leading to alternative exit sites E1 and E2, respectively. Next: The X-ray Structure Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture22/lecture3_4.html (6 of 6)5/24/2007 12:51:37 PM The X-ray Structure of the Ribosome (3.4b) The X-ray Structure of the Ribosome Two back-to-back landmark papers in the journal Science (August 2000) provided a whole new level of information of what ribosomes look like in detail, and specifically, how the crucial peptide bond formation is catalyzed: The Complete Atomic Structure of the Large Ribosomal Subunit at 2.4 Å Resolution Nenad Ban, Poul Nissen, Jeffrey Hansen, Peter B. Moore, Thomas A. Steitz The large ribosomal subunit catalyzes peptide bond formation and binds initiation, termination, and elongation factors. We have determined the crystal structure of the large ribosomal subunit from Haloarcula marismortui at 2.4 angstrom resolution, and it includes 2833 of the subunit's 3045 nucleotides and 27 of its 31 proteins. The domains of its RNAs all have irregular shapes and fit together in the ribosome like the pieces of a three-dimensional jigsaw puzzle to form a large, monolithic structure. Proteins are abundant everywhere on its surface except in the active site where peptide bond formation occurs and where it contacts the small subunit. Most of the proteins stabilize the structure by interacting with several RNA domains, often using idiosyncratically folded extensions that reach into the subunit's interior. Science (2000) 289, 905-920. The Structural Basis of Ribosome Activity in Peptide Bond Synthesis Poul Nissen, Jeffrey Hansen, Nenad Ban, Peter B. Moore, Thomas A. Steitz Using the atomic structures of the large ribosomal subunit from Haloarcula marismortui and its complexes with two substrate analogs, we establish that the ribosome is a ribozyme and address the catalytic properties of its all-RNA active site. Both substrate analogs are contacted exclusively by conserved ribosomal RNA (rRNA) residues from domain V of 23S rRNA; there are no protein side-chain atoms closer than about 18 angstroms to the peptide bond being synthesized. The mechanism of peptide bond synthesis appears to resemble the reverse of the acylation step in serine proteases, with the base of A2486 (A2451 in Escherichia coli) playing the same general base role as histidine-57 in chymotrypsin. The unusual pKa (where Ka is the acid dissociation constant) required for A2486 to perform this function may derive in part from its hydrogen bonding to G2482 (G2447 in E. coli), which also interacts with a buried phosphate that could stabilize unusual tautomers of these two bases. The polypeptide exit tunnel is largely formed by RNA but has significant contributions from proteins L4, L22, and L39e, and its exit is encircled by proteins L19, L22, L23, L24, L29, and L31e. Science (2000) 289, 920-930. http://bass.bio.uci.edu/~hudel/bs99a/lecture22/lecture3_4b.html (1 of 6)5/24/2007 12:51:49 PM The X-ray Structure of the Ribosome (3.4b) The first paper provides a wealth of structural information on how the components of the large (50S) subunit are arranged: http://bass.bio.uci.edu/~hudel/bs99a/lecture22/lecture3_4b.html (2 of 6)5/24/2007 12:51:49 PM The X-ray Structure of the Ribosome (3.4b) ● ● the rRNA (in gray) is forming the core the ribosomal proteins (yellow) are mostly on the surface It also shows the exit tunnel and suggests that not only an extended polpypeptide would fit through it, but also one in an alpha-helical conformation: http://bass.bio.uci.edu/~hudel/bs99a/lecture22/lecture3_4b.html (3 of 6)5/24/2007 12:51:49 PM The X-ray Structure of the Ribosome (3.4b) The second paper provides a detailed mechanism for peptide bond formation based on the x-ray structure. It http://bass.bio.uci.edu/~hudel/bs99a/lecture22/lecture3_4b.html (4 of 6)5/24/2007 12:51:49 PM The X-ray Structure of the Ribosome (3.4b) highlights the central role of the rRNA, and specifically that of base A2486 (RIBOZYME!): Next: Supplemental Material http://bass.bio.uci.edu/~hudel/bs99a/lecture22/lecture3_4b.html (5 of 6)5/24/2007 12:51:49 PM The X-ray Structure of the Ribosome (3.4b) Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture22/lecture3_4b.html (6 of 6)5/24/2007 12:51:49 PM Supplemental Material (3.5) Supplemental Material This part won't be on the final! If you are interested how findings like these are presented in an original research article, you should take a look at the 1995 paper in the journal Nature: "A model of protein synthesis based on cryo-electron microscopy of the E. coli ribosome" by Frank, J., Zhu, J., Penczek, P., Li, Y., Srivastava, S., Verschoor, A., Radermacher, M., Grassucci, R., Lata, R.K. and Agrawal, R.K. Nature 376, 441-444 (1995). And for comparison, check out the more recent back-to-back X-ray papers in the journal Science: "The Complete Atomic Structure of the Large Ribosomal Subunit at 2.4 Å Resolution" by Nenad Ban, Poul Nissen, Jeffrey Hansen, Peter B. Moore, Thomas A. Steitz Science 289, 905-920 (2000). http://bass.bio.uci.edu/~hudel/bs99a/lecture22/lecture3_5.html (1 of 2)5/24/2007 12:51:56 PM Supplemental Material (3.5) "The Structural Basis of Ribosome Activity in Peptide Bond Synthesis" by Poul Nissen, Jeffrey Hansen, Nenad Ban, Peter B. Moore, Thomas A. Steitz. Science 289, 920-930 (2000). These issues are available online or as hardcopies at the UCI Science Library. Next: Summary Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture22/lecture3_5.html (2 of 2)5/24/2007 12:51:56 PM Summary (3.6) Summary ● ● ● ● ● ● ● ● Ribosomes are large and complex molecular precision machines. Ribosomes occur free in the cytosol or attached to the endoplasmic reticulum. They contain two subunits, called small and large. Both subunits are comprised of large rRNAs and many proteins. rRNAs form the core and are stabilized by extended base-stacking & base-pairing. Most ribosomal proteins are located on the surface. Peptide-bond formation is catalyzed one base of the 23S rRNA (ribozyme) in the large subunit. Ribosomes have tunnels, channels and cavities which accommodate the various players during translation: mRNA, aminoacyl-tRNA, nascent protein. Next Lecture: Initiation of Protein Synthesis Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture22/lecture3_6.html5/24/2007 12:52:04 PM Initiation of Protein Synthesis (4.1) Lecture 4 Polypeptide Synthesis Overview Polypeptide synthesis proceeds sequentially from N Terminus to C terminus. Amino acids are not pre-positioned on a template. Proved by the classic experiment of Dintzis & coworkers in 1961. They synthesized hemoglobin in a test tube using a reticulocyte (red blood cell) system. They initiated protein synthesis and then added H3labeled leucine. They isolated hemoglobin, partially labeled with incorporated H3-leucine, and created peptide fragments (labeled a, b, c, d, e, f, g below) by digestion of hemoglobin with trypsin. Then they analyzed peptide fragments to determine the relative amount of radioactivity. The following results were obtained: http://bass.bio.uci.edu/~hudel/bs99a/lecture23/index.html (1 of 5)5/24/2007 12:52:28 PM Initiation of Protein Synthesis (4.1) http://bass.bio.uci.edu/~hudel/bs99a/lecture23/index.html (2 of 5)5/24/2007 12:52:28 PM Initiation of Protein Synthesis (4.1) Figure 4.1.1: Incorporation of radioactive leucine into hemoglobin after different periods of incubation. There is a gradient from the C-terminus to the N-terminus of the protein, which implies that synthesis takes place sequentially from N-terminus to C-terminus. As the time of incubation is increased, the total 3 amount of radioactivity is increased but the gradient remains. Results are expressed as the ratio of an H 14 label (tritium) in leucine to an internal control of C . Data of Dintzis (1961). http://bass.bio.uci.edu/~hudel/bs99a/lecture23/index.html (3 of 5)5/24/2007 12:52:28 PM Initiation of Protein Synthesis (4.1) Ribosomes read mRNA in the 5'->3' direction, not the 3'->5' direction. To prove, one must know the Genetic Code. For example, AAA codes for Lys, AAC codes for Asn and CAA codes for Gln. In a cell-free protein synthesizing system, initiate protein synthesis by adding artificial mRNA, consisting of polyA with one C (cytosine) at the 3' end. Analyze the N and C termini of the peptide fragments. Nucleotide Sequence Protein Sequence Direction of Synthesis 5'-AAAAAA(AAA)nAAC-3' Lys-Lys-(Lys)n-Asn 5' -> 3' 3'-CAAAAA(AAA)nAAA-5' Gln-Lys-(Lys)n-Lys 3' -> 5' Next: Polypeptide Synthesis Overview (continued) http://bass.bio.uci.edu/~hudel/bs99a/lecture23/index.html (4 of 5)5/24/2007 12:52:28 PM Initiation of Protein Synthesis (4.1) Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture23/index.html (5 of 5)5/24/2007 12:52:28 PM Initiation of Protein Synthesis (4.2) Polypeptide Synthesis Overview (cont.) Active translation occurs on polyribosomes Electron micrographs of ribosomes actively engaged in protein synthesis revealed by "beads on a string" appearance. This implied that each mRNA transcript is read simultaneously by more than one ribosome. In fact, a second, third, fourth, etc. ribosome starts to read the mRNA transcript before the first ribosome has completed the synthesis of one polypeptide chain. Multiple ribosomes on a single mRNA transcript are called polyribosomes or polysomes. The individual ribosomes are separated by gaps of 50 Å to 150 Å. Thus, there is approximately one ribosome per 80 nucleotides. http://bass.bio.uci.edu/~hudel/bs99a/lecture23/lecture4_2.html (1 of 4)5/24/2007 12:52:37 PM Initiation of Protein Synthesis (4.2) Chain elongation links the growing polypeptide to incoming aminoacyl-tRNA The newly formed polypeptide chain is attached to a tRNA molecule, not to a protein or mRNA molecule. To prove, use a cell-free protein synthesizing system with artificial mRNA, such as polyA. Add H3-Lys. Wash active ribosomes in high salt to dissociate and stop protein synthesis. Analyze results and find highest concentration of H3-Lys -- always with tRNA. From many such experiments, there are now known to be three tRNA binding sites on the ribosomes: A Site The ribosomal site which binds the incoming aminoacyl-tRNA (Acceptor site). P Site The site which holds the peptidyl-tRNA, that is the tRNA which is covalently linked to the growing polypeptide chain (Peptidyl site). E Site A site which transiently binds to the outgoing, deacylated tRNA (Exit site). http://bass.bio.uci.edu/~hudel/bs99a/lecture23/lecture4_2.html (2 of 4)5/24/2007 12:52:37 PM Initiation of Protein Synthesis (4.2) Three steps are involved in the synthesis of one polypeptide chain 1. INITIATION ❍ ❍ Assembly of active ribosome and the reading of the first mRNA codon (START codon). Occurs once per polypeptide chain. 2. ELONGATION ❍ Involves three distinct steps for each mRNA codon or amino acid in the new http://bass.bio.uci.edu/~hudel/bs99a/lecture23/lecture4_2.html (3 of 4)5/24/2007 12:52:37 PM Initiation of Protein Synthesis (4.2) polypeptide chain: 1. Transfer of aminoacyl-tRNA from cytoplasm to A-site of ribosome. 2. Covalent linkage of new amino acid to growing polypeptide chain peptidyl transfer. 3. Movement of tRNA from A-site to P-site and simultaneous movement of mRNA by 3 nucleotides - translocation. ❍ Occurs multiple times per polypeptide. 3. TERMINATION ❍ ❍ Reading of final mRNA codon (STOP codon) and dissociation of polypeptide from ribosome. Occurs once per polypeptide chain. Next: Initiation of Protein Synthesis Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture23/lecture4_2.html (4 of 4)5/24/2007 12:52:37 PM Initiation of Protein Synthesis (4.3) Initiation of Protein Synthesis INITIATION - Assembly of active ribosome and the reading of the first mRNA codon. Occurs once per polypeptide chain. The signals for initiation: mRNA Start Codon Prokaryotes: Eukaryotes: AUG (very rarely also GUG & UUG) AUG After protein is made, the N-terminal methionine is frequently cleaved off. Shine-Dalgarno Sequence To distinguish the start AUG codon from AUG codons which code for internal methionines or from a fortuitous AUG combination in another reading frame, the start AUG codon in prokaryotes is preceded by a highly conserved sequence at the 5´ end of the mRNA transcript which serves as a ribosome-binding site. The Shine-Dalgarno sequence is a purine-rich tract of 3-10 nucleotides and precedes the start AUG codon by approximately 10 nucleotides upstream on the 5´ side. The Shine-Dalgarno sequence forms base pairing with a highly conserved, pyrimidine-rich region of the 16S rRNA http://bass.bio.uci.edu/~hudel/bs99a/lecture23/lecture4_3.html (1 of 4)5/24/2007 12:52:43 PM Initiation of Protein Synthesis (4.3) which is part of the 30S ribosomal subunit. This base-pairing properly aligns the start AUG codon in the P site of the 30S ribosomal subunit. In eukaryotes, protein synthesis usually begins with the first AUG codon from the 5´ end. Eukaryotic mRNA has a special cap at the 5´ end of the mRNA which is recognized by a cap-binding protein. Special Initiator tRNA Only one special tRNA is used to place the first amino acid, which is always a formyl-methionine (fMet): http://bass.bio.uci.edu/~hudel/bs99a/lecture23/lecture4_3.html (2 of 4)5/24/2007 12:52:43 PM Initiation of Protein Synthesis (4.3) The initiator tRNA is called fMet-tRNA. It differs from all other tRNAs by one less base pair in the acceptor stem region and by three consecutive G:C base pairs in the anticodon region. These changes alter the 3-dimensional structure of the fMet-tRNA in subtle ways. fMet-tRNAfMet and Met-tRNAMet both recognize the AUG codon. A methionine is attached to both tRNAs by the same methionyl tRNA synthetase. However, once the methionine is attached to either tRNA, a special enzyme called formyl transferase, which recognizes only the initiator tRNAfMet adds a formyl group to the amide nitrogen. Because the amide N is now blocked from further reaction, this amino acid can only be positioned at the start of a polypeptide chain. Soluble Protein Factors In prokaryotes, there are three initiation factors: IF1, IF2, IF3, named in order of discovery, not in order of their function on the ribosome. The initiation factors are proteins that facilitate the assembly of an active http://bass.bio.uci.edu/~hudel/bs99a/lecture23/lecture4_3.html (3 of 4)5/24/2007 12:52:43 PM Initiation of Protein Synthesis (4.3) ribosomal particle and the placement of the first fMet-tRNAfMet. In eukaryotes, there are 10 initiation factors, called eIF1, eIF2, etc. IF3 binds to 30S subunit and promotes the dissociation of the 30S subunit from the 50S subunit and the subsequent binding of the mRNA. IF1 helps IF3 by increasing the dissociation rate between the 30S and 50S subunit. IF2, in complex with GTP, binds to fMet-tRNAfMet and places it in the P-site of the 30S subunit. Then IF3 is released, causing GTP to hydrolyze to GDP, which in turn releases IF2-GDP. GTP hydrolysis promotes the release of IF1 and the subsequent association of the 30S subunit with the 50S subunit. Next: Schematic of Initiation Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture23/lecture4_3.html (4 of 4)5/24/2007 12:52:43 PM Initiation of Protein Synthesis in Prokaryotes (4.4) Formation of the 70S initiation complex Schematic of the initiation of protein synthesis in prokaryotes http://bass.bio.uci.edu/~hudel/bs99a/lecture23/lecture4_4.html (1 of 3)5/24/2007 12:52:49 PM Initiation of Protein Synthesis in Prokaryotes (4.4) 1. Binding on IF1 & IF3 to an empty 70S ribosome dissociates it into 50S and 30S subunits. 2. a. 5' region of mRNA binds to free 30S ribsomal subunit, IF3 is released. b. Initiator fMet-tRNAfMet carried by IF2-GTP binds to P site of 30S ribsomal subunit. 3. Free 50S ribosomal subunit binds, IF1 & IF2-GDP + Pi are released. Certain details of the process remain uncertain. For example, the exact order of binding of IF1, IF2, IF3, tRNA, and mRNA is unclear. The above is one current model (from the text book). Next: Initiation of translation in eukaryotes http://bass.bio.uci.edu/~hudel/bs99a/lecture23/lecture4_4.html (2 of 3)5/24/2007 12:52:49 PM Initiation of Protein Synthesis in Prokaryotes (4.4) Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture23/lecture4_4.html (3 of 3)5/24/2007 12:52:49 PM Initiation of Protein Synthesis in Eukaryotes (4.5) Schematic of the initiation of protein synthesis in eukaryotes http://bass.bio.uci.edu/~hudel/bs99a/lecture23/lecture4_5.html (1 of 2)5/24/2007 12:52:55 PM Initiation of Protein Synthesis in Eukaryotes (4.5) Note that the process is in principle very similar to prokaryotic initiation. Major differences from prokaryotic initiation are associated with the m7G cap of the mRNA and its interaction with the cap-binding protein (CBPI) and eIF4F (CBPII) instead of Shine-Dalgarno sequencemediated alignment to properly position the first AUG codon in the P site of the ribosome. Next: Summary Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture23/lecture4_5.html (2 of 2)5/24/2007 12:52:55 PM Summary (4.6) Summary ● ● ● ● ● Polypeptide synthesis proceeds sequentially from N terminus to C terminus Ribosomes read mRNA in the 5'->3' direction, not the 3'->5' direction Active translation typically occurs on polyribosomes Chain elongation links the growing polypeptide to incoming aminoacyl-tRNA Three steps are involved in the synthesis of one polypeptide chain of N residues: 1. INITIATION (1 x) 2. ELONGATION (N-1 x) 3. TERMINATION (1 x) ● Initiation in prokaryotes requires: 1. One empty 70S ribosome 2. Three initiation factors: IF1, IF2, IF3 3. mRNA with Shine-Dalgarno sequence and START codon (AUG) 4. Initiator fMet-tRNAfMet 5. One GTP ● Initiation in eukaryotes is similar, but more complicated Next lecture: Elongation & Termination of Protein Synthesis http://bass.bio.uci.edu/~hudel/bs99a/lecture23/lecture4_6.html (1 of 2)5/24/2007 12:53:00 PM Summary (4.6) Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture23/lecture4_6.html (2 of 2)5/24/2007 12:53:00 PM Elongation & Termination of Protein Synthesis (5.1) Lecture 5 1. INITIATION ● ● Assembly of active ribosome by placing the first mRNA codon (AUG or START codon) near the P site and pairing it with initiation tRNA, fMet-tRNAfMet. Occurs once per polypeptide chain or molecule. 2. ELONGATION ● Involves three distinct steps for each mRNA codon or amino acid in the new polypeptide chain: 1. Transfer of proper aminoacyl-tRNA from cytoplasm to A-site of ribosome. 2. Covalent linkage of new amino acid to growing polypeptide chain - peptidyl transfer. 3. Movement of tRNA from A-site to P-site and simultaneous movement of mRNA by 3 nucleotides - translocation. ● Occurs multiple times per polypeptide. 3. TERMINATION ● ● Reading of final mRNA codon (STOP codon) and dissociation of polypeptide from ribosome. Occurs once per polypeptide chain. http://bass.bio.uci.edu/~hudel/bs99a/lecture24/index.html (1 of 5)5/24/2007 12:53:05 PM Elongation & Termination of Protein Synthesis (5.1) ELONGATION OF PROTEIN SYNTHESIS ● ● ● ● ● 3 distinct steps to add one amino acid to the growing polypeptide chain. Occurs many times per polypeptide, the number of which depends upon the number of mRNA codons or amino acids in the protein The Elongation Cycle is similar in prokaryotes and eukaryotes. Fast: 15-20 amino acids added per second Accurate: 1 mistake every ~10,000 amino acids Transfer of Aminoacyl-tRNA Non-initiator, aminoacyl-tRNA is placed in the ribosomal A-site over the mRNA codon in such a way that base pairing occurs between the anticodon loop of the tRNA and mRNA codon (see lecture 1). Aminoacyl-tRNA transfer is facilitated by two soluble protein transfer factors, called elongation factors, EF-Tu and EF-Ts in prokaryotes: ❍ ❍ EF-Tu binds GDP and GTP and is a model G protein. EF-Ts exchanges GDP for GTP on EF-Tu. The elongation factors are similar in eukaryotes. Instead of two proteins, there is a stable trimer, eEF1-alpha-beta-gamma, which carries out the same function as EF-Tu and EF-Ts. eEF1-alpha is the eukaryotic equivalent of EF-Tu, and eEF1-beta-gamma the eukaryotic equivalent of EF-Ts. EF-Tu-GDP is the inactive form. EF-Ts activates EF-Tu by catalyzing the exchange of GDP for GTP. EF-Tu-GTP is the active form which binds to non-initiator tRNAs to which the aminoacyl http://bass.bio.uci.edu/~hudel/bs99a/lecture24/index.html (2 of 5)5/24/2007 12:53:05 PM Elongation & Termination of Protein Synthesis (5.1) group has been attached. EF-Tu-GTP-aminoacyl-tRNA is then carried to the ribosome. The complex binds to the ribosome, with the aminoacyl-tRNA in the A-site. Ribosome binding stimulates GTP hydrolysis and EF-Tu-GDP dissociates from the ribosome, free to recycle through the step multiple times. http://bass.bio.uci.edu/~hudel/bs99a/lecture24/index.html (3 of 5)5/24/2007 12:53:05 PM Elongation & Termination of Protein Synthesis (5.1) EF-Tu is partially responsible for the high degree of accuracy of protein synthesis via a mechanism called kinetic proofreading. The error rate of protein synthesis is only 1 wrong amino acid for every 10,000 amino acids added to polypeptides. When a charged tRNA is positioned into the A-site, the anticodon must base pair with the mRNA codon. If there is an incorrect match, the incorrect aa-tRNA dissociates from the ribosome before GTP hydrolysis occurs. If there is a correct match, GTP hydrolysis occurs and EF-Tu-GDP leaves the ribosome before the cognate aminoacyl-tRNA can dissociate and EF-Tu-GDP dissociates instead, leaving the correct tRNA on the ribosome. EF-Tu is so important to cellular function that it is one of the most abundant cytoplasmic proteins (>5%). There is one copy of the EF-Tu protein for each tRNA molecule in the cell. Next: Peptidyl Transfer http://bass.bio.uci.edu/~hudel/bs99a/lecture24/index.html (4 of 5)5/24/2007 12:53:05 PM Elongation & Termination of Protein Synthesis (5.1) Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture24/index.html (5 of 5)5/24/2007 12:53:05 PM Elongation & Termination of Protein Synthesis (5.2) ELONGATION OF PROTEIN SYNTHESIS (cont.) Peptidyl Transfer or Transpeptidation Next, the growing polypeptide chain on the P-site tRNA (peptidyl tRNA) is covalently linked to the amino acid attached to the A-site tRNA, that is a peptide bond is formed. The P-site tRNA is now unacylated and the A-site tRNA is covalently linked to the growing polypeptide chain. This step is somewhat of a mystery but is believed to be catalyzed by a region of the 50S subunit, called the peptidyltransferase complex. The complex utilizes both proteins and 23S rRNA. There is growing evidence that 23S rRNA actively catalyzes the peptidyl transfer step but the mode of action is yet unknown. Translocation The A-site tRNA with the growing polypeptide chain is then moved within the ribosome to the P-site while the deacylated P-site tRNA is moved to the E-site. Simultaneously, the mRNA is shifted by exactly 3 ribonucleotides or 1 codon. http://bass.bio.uci.edu/~hudel/bs99a/lecture24/lecture5_2.html (1 of 2)5/24/2007 12:53:16 PM Elongation & Termination of Protein Synthesis (5.2) The process requires a soluble protein factor, called elongation factor EF-G. EF-G is similar, but much larger than EFTu. EF-G is also active as the GTP complex. Although many older texts report that the binding of EF-G-GTP to the ribosome provides the energy for translocation, it is now known to be wrong. The energy derived from GTP hydrolysis drives translocation. After GTP hydrolysis, EF-G-GDP is released from the ribosome, the tRNA carrying the polypeptide chain is in the P site, and the next mRNA codon is in the A site. The ribosome is now ready to start the elongation cycle over again to add a new amino acid, until a termination signal is reached. It appears that EF-G catalyzes its own exchange of GDP and GTP. No soluble guanine nucleotide exchange factor has yet been found. The equivalent of EF-G in eukaryotes is eEF2. Next: Schematic of Elongation Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture24/lecture5_2.html (2 of 2)5/24/2007 12:53:16 PM Elongation & Termination of Protein Synthesis (5.3) Chain elongation in prokaryotic translation http://bass.bio.uci.edu/~hudel/bs99a/lecture24/lecture5_3.html (1 of 2)5/24/2007 12:53:23 PM Elongation & Termination of Protein Synthesis (5.3) The elongation process is depicted as a cycle. Following translocation (step 3) and empty tRNA release (step 4), the ribosome is ready to accept the next aminoacyl tRNA (aa-tRNA) and repeat the cycle. This cycle will continue until a termination codon is reached. Next: Atomic Structures of EF-Tu Complexes Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture24/lecture5_3.html (2 of 2)5/24/2007 12:53:23 PM Elongation & Termination of Protein Synthesis (5.4) ATOMIC STRUCTURES OF EF-Tu COMPLEXES http://bass.bio.uci.edu/~hudel/bs99a/lecture24/lecture5_4.html (1 of 2)5/24/2007 12:54:15 PM Elongation & Termination of Protein Synthesis (5.4) Next: Termination of Protein Synthesis Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture24/lecture5_4.html (2 of 2)5/24/2007 12:54:15 PM Elongation & Termination of Protein Synthesis (5.5) TERMINATION OF PROTEIN SYNTHESIS 1. The mRNA Signal STOP Codons: UAA, UAG, or UGA There are no tRNAs that recognize the STOP codons UAA, UAG, or UGA. 2. Soluble Protein Release Factors RF1 responds to UAA or UAG RF2 responds to UAA or UGA RF3, a GTPase (like EF-Tu and binds in a similar A-site location) RF1/RF2 interact with RF3-GTP, have a similar shape as EF-Tu-GTP-aa-tRNA or EF-G, and bind in a similar ribosomal site (A-site). In a manner similar to EF-G, GTP hydrolysis drives the movement of the terminal mRNA codon into the P-site, moving the last tRNA into the E-site and off. At the same time, the polypeptide chain is released after hydrolysis of the tRNA-peptide bond. In eukaryotes, only a single release factor, eRF, is necessary. It recognizes all three STOP codons and interacts with GTP. A mutation resulting in a premature STOP codon is called a nonsense mutation. http://bass.bio.uci.edu/~hudel/bs99a/lecture24/lecture5_5.html (1 of 3)5/24/2007 12:54:20 PM Elongation & Termination of Protein Synthesis (5.5) http://bass.bio.uci.edu/~hudel/bs99a/lecture24/lecture5_5.html (2 of 3)5/24/2007 12:54:20 PM Elongation & Termination of Protein Synthesis (5.5) The termination pathway in E. coli ribosomes: RF-1 recognizes the termination codons UAA and UAG, whereas RF-2 recognizes UAA and UGA. Eukaryotic termination follows an analogous pathway but requires only a single release factor, eRF, that recognizes all three termination codons. Next: Summary Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture24/lecture5_5.html (3 of 3)5/24/2007 12:54:20 PM Summary (5.6) Summary ● ● ● ● ● ● ● ● ● ● ● ● Elongation consists of three distinct steps to add one amino acid Requires three elongation factors: EF-Tu/EF-Ts and EF-G Requires two GTPs per cycle Occurs many times per polypeptide The elongation cycle is similar in prokaryotes and eukaryotes. Fast: 15-20 amino acids added per second Accurate: 1 mistake every ~10,000 amino acids Termination results in the release of the polypeptide chain Requires one of the three STOP codons: UAA, UAG, or UGA. Requires RF1 or RF2, and RF3 in prokaryotes (eRF in eukaryotes) Requires one GTP Each step of protein synthesis (initiation, elongation and termination) requires GTP Next lecture: Regulation of Protein Synthesis at the Translational Level And now: Movie time! http://bass.bio.uci.edu/~hudel/bs99a/lecture24/lecture5_6.html (1 of 2)5/24/2007 12:54:25 PM Summary (5.6) Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture24/lecture5_6.html (2 of 2)5/24/2007 12:54:25 PM Regulation of Protein Synthesis (6.1) Lecture 6 Regulation of Protein Synthesis at the Translational Level Comparison of EF-Tu-GDP and EF-Tu-GTP conformations EF-Tu-GDP EF-Tu-GTP Next: Comparison of GDP and GTP binding region in EF-Tu Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture25/index.html5/24/2007 12:54:33 PM Regulation of Protein Synthesis (6.1a) Comparison of GDP and GTP binding region of EF-Tu http://bass.bio.uci.edu/~hudel/bs99a/lecture25/lecture6_1a.html (1 of 2)5/24/2007 12:54:38 PM Regulation of Protein Synthesis (6.1a) Next: Molecular Mimicry Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture25/lecture6_1a.html (2 of 2)5/24/2007 12:54:38 PM Regulation of Protein Synthesis (6.1b) Molecular mimicry between EF-G and EF-Tu-GTP-aminoacyl-tRNA EF-G EF-Tu-GTP-Aminoacyl-tRNA Next: Rates and Energetics of Translation Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture25/lecture6_1b.html5/24/2007 12:54:44 PM Regulation of Protein Synthesis (6.2) Rates and energetics of translation At 37° C, the rate of translation in E. coli is about 15 amino acids per second. The translational rate is equivalent to the transcriptional rate which is ~45 nucleotides per second. Energy cost for synthesis of a protein with N amino acids: 2N 1 ATPs to charge tRNA (ATP -> AMP + PP -> AMP + 2Pi) GTP for initiation (IF2) N-1 GTPs to position tRNA for N-1 peptide bonds (EF-Tu) N-1 GTPs for N-1 translocation steps (EF-G) 1 GTP for termination (RF-3) ==== 4N Total of 4 high-energy phosphate bonds cleaved per amino acid Each ATP or GTP cleavage generates ~40 kJ/mol Each peptide bond costs ~160 kJ/mol in the cell, yet an uncatalyzed chemical reaction to form a peptide bond costs only ~20 kJ/mol. http://bass.bio.uci.edu/~hudel/bs99a/lecture25/lecture6_2.html (1 of 7)5/24/2007 12:54:49 PM Regulation of Protein Synthesis (6.2) Why is it so costly to make a peptide bond on a ribosome? The excess energy is used for generating an accurate, defined polypeptide sequence, not a random one or a combination of multiple possibilities. Two sources of errors during translation: ● ● Attachment of an incorrect amino acid to a tRNA Mispairing of the tRNA anticodon with the mRNA codon Two proofreading mechanisms exist to prevent these errors: ● ● Proofreading before aminoacyl adenylate intermediate is attached to tRNA. Kinetic proofreading before peptide bond formation: A delay is introduced between the binding of an aminoacyl-tRNA to the codon and the formation of the peptide bond to allow errors to be corrected: EF-Tu-GTP binds an aminoacyl-tRNA and bring it into the A-site. EF-Tu allows the anticodon to interact with the codon but prevents peptide bond formation. An incorrect tRNA will bind weakly to the codon and will dissociate from the codon before an incorrect amino acid is incorporated into the polypeptide. Correct codon-anticodon matching triggers hydrolysis of GTP by the EF-Tu, after which EF-Tu-GDP dissociates. Peptide bond formation proceeds. ❍ ❍ ❍ ❍ ❍ http://bass.bio.uci.edu/~hudel/bs99a/lecture25/lecture6_2.html (2 of 7)5/24/2007 12:54:49 PM Regulation of Protein Synthesis (6.2) Each major step in protein synthesis, except peptide-bond formation itself, involves hydrolysis of GTP to GDP. Regulation of protein synthesis PROKARYOTES Short-lived mRNA (few minutes), so little need for complicated translational regulation. In prokaryotes, most of the regulation is at the transcriptional level. Rates vary only by a factor of 100. Variance is due to differences in ShineDalgarno sequences and how strongly a particular sequence base-pairs with the 16S rRNA of the 30S ribosomal subunit. EUKARYOTES Long-lived mRNA (hours to days) and thus a greater need to regulate the rate of protein synthesis. http://bass.bio.uci.edu/~hudel/bs99a/lecture25/lecture6_2.html (3 of 7)5/24/2007 12:54:49 PM Regulation of Protein Synthesis (6.2) Several known mechanisms: ● ● mRNA masking: mRNA is bound to a variety of proteins that prevent association with ribosomes. When appropriate signal is received, the proteins dissociate from mRNA, leaving the transcript free to associate with the ribosome. The signal is usually in the form of phosphorylation/ dephosphorylation. mRNA masking is a major form of regulation in early embryonic development. antisense RNA: short segment of RNA, complementary to mRNA, that forms double stranded RNA which cannot be translated by ribosome. Two known examples: ❍ ❍ ● blockage of protein synthesis of fruit-ripening enzyme in tomatoes the c-myb gene product which promotes smooth muscle development and blockage in injured arteries. Heme Control of Globin Synthesis: Red blood cells are programmed to synthesize large amounts of globin. The globin chains, subsequent to translation, are assembled with heme into hemoglobin. If there is an insufficient supply of heme to insert into the newly synthesized globin chains, then translation is turned off. The lack of heme triggers the accumulation of a heme-controlled inhibitor (HCI) protein. This protein is a kinase which phosphorylates eIF2-GTP. The phosphorylation blocks the dissociation of eIF2 and eIF2-beta that normally occurs in the initiation cycle. Thus, the cell becomes rapidly depleted of unphosphorylated eIF2 which is normally recycled for initiation of additonal mRNA. Either the http://bass.bio.uci.edu/~hudel/bs99a/lecture25/lecture6_2.html (4 of 7)5/24/2007 12:54:49 PM Regulation of Protein Synthesis (6.2) addition of heme, which represses the production of HCI, or the addition of lots of unphosphorylated eIF2, which bypasses the HCI effect, can restart initiation again. ● Interferon: Interferons are glycoproteins that are secreted by virusinfected cells. Interferons prevent additional infection by other types of viruses by inhibiting protein synthesis in infected cells. Two mechanisms of action: ❍ ❍ induces production of protein kinase, DAI (double-stranded RNAactivated inhibitor) which in the presence of dsRNA, phosphorylates eIF2-alpha and stabilizes the eIF2-alpha-eIF2-beta complex in a manner similar to the heme-controlled inhibitor (HCI). induces a cascade effect which ultimately activates an endonuclease, RNase L, that rapidly degrades mRNA. Inhibition of protein synthesis by antibiotics http://bass.bio.uci.edu/~hudel/bs99a/lecture25/lecture6_2.html (5 of 7)5/24/2007 12:54:49 PM Regulation of Protein Synthesis (6.2) Antibiotics are bacterially or fungally produced substances that inhibit the growth of other organisms. Antibiotics target a wide spectrum of vital processes: they block DNA replication, transcription and bacterial cell wall synthesis. A large number of antibiotics, including medically useful substances, block protein translation. Blocking protein translation is very effective for two reasons: ● ● Protein translation plays a central role in overall metabolism The structural differences between prokaryotic and eukaroytic ribosomes and associated factors (IFs/EFs/RFs) allow specific targeting. http://bass.bio.uci.edu/~hudel/bs99a/lecture25/lecture6_2.html (6 of 7)5/24/2007 12:54:49 PM Regulation of Protein Synthesis (6.2) Prokaryotic Inhibitors ● ● ● ● ● ● Chloramphenicol - inhibits peptidyl transferase on 50S subunit. Erythromycin - inhibits translocation by 50S subunit. Fusidic acid - inhibits translocation by preventing the dissociation of EF-GGDP from ribosome. Puromycin - an aminoacyl-tRNA analog that causes premature chain termination. Streptomycin - causes mRNA misreading and inhibits chain initiation. Tetracycline - inhibits binding of aminoacyl-tRNA to ribosomal A-site. Eukaryotic Inhibitors ● ● ● Puromycin & Tetracycline (see prokaryotes above). Cycloheximide - inhibits peptidyl transferase on 60S subunit. Diphtheria Toxin - inactivates eEF-2 by ADP ribosylation. Summary Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture25/lecture6_2.html (7 of 7)5/24/2007 12:54:49 PM http://bass.bio.uci.edu/~hudel/bs99a/lecture21/trna_charge2.gif http://bass.bio.uci.edu/~hudel/bs99a/lecture21/trna_charge2.gif5/24/2007 12:55:02 PM Summary (6.3) Summary ● ● ● ● ● ● ● ● GTP-binding often causes major conformational changes in proteins Each major step in protein synthesis, except peptide-bond formation itself, involves hydrolysis of GTP to GDP. Four high-energy phosphate-bonds are required for each amino acid added The rate of protein synthesis is well matched to the rate of transcription EF-G is shaped like EF-Tu-GTP-aminoacyl-tRNA (molecular mimicry) mRNA in prokaryotes is short-lived (minutes) mRNA in eukaryotes is long-lived (hours/days), requiring additional control A large number of antibiotics inhibit protein synthesis, many specifically in prokaryotes Next lecture: Post-Translational Processing Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture25/lecture6_3.html5/24/2007 12:55:23 PM Post-Translational Processing (7.1) Lecture 7 Post-Translational Processing Protein synthesis consists of several steps: from the translation of the information from mRNA to the folded and fully processed, active protein in its proper compartment of action. The mRNA sequence predicts a specific length polypeptide chain made up of the primary 20 amino acids. Fully processed protein products are almost always shorter than their mRNA would predict, and globally contain about 200 different amino acids. http://bass.bio.uci.edu/~hudel/bs99a/lecture26/index.html (1 of 4)5/24/2007 12:55:29 PM Post-Translational Processing (7.1) (determined by sequencing, biochemistry, X-ray crystallography) During translation, about 30-40 polypeptide residues are relatively protected by the ribosome (tunnel T and exit sites E1 and E2 in the large subunit). Once the polypeptide chain emerges from the ribosome it starts to fold and can be subject to post-translational modifications. http://bass.bio.uci.edu/~hudel/bs99a/lecture26/index.html (2 of 4)5/24/2007 12:55:29 PM Post-Translational Processing (7.1) So after translation several additional steps must be considered as part of the complete protein biosynthetic process: 1. Covalent modification of a: b: c: d: peptide bonds the N-terminus the C-terminus amino acid residues (side chains). 2. Noncovalent modifications: folding, addition of co-factors. 3. Translocation: compartment selection and transport (Trafficking/Targeting). 4. Involvement of molecular chaperones in 1, 2, and 3. Why post-translational processing? ● adds functionality http://bass.bio.uci.edu/~hudel/bs99a/lecture26/index.html (3 of 4)5/24/2007 12:55:29 PM Post-Translational Processing (7.1) ● ● ● ● effects targeting regulates activity increases mechanical strength changes recognition Next: Covalent Modifications Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture26/index.html (4 of 4)5/24/2007 12:55:29 PM Covalent Modifications (7.2) Covalent Modifications Modifications involving the peptide bond (peptide bond cleavage or limited proteolysis): ● usually carried out by enzymes called peptidases or proteases: ❍ ❍ ❍ ❍ activation of proenzymes (digestive enzymes, blood clotting cascade, complement activation etc.) and prohormones (insulin) production of active neuropeptides and peptide hormones from high molecular weight precursors macromolecular assembly in virus particles (e.g. HIV protease) removal of signal sequences http://bass.bio.uci.edu/~hudel/bs99a/lecture26/lecture7_2.html (1 of 4)5/24/2007 12:55:35 PM Covalent Modifications (7.2) These reactions are often exquisitely specific for only one or a few peptide bonds. Modifications involving the amino terminus: ● ● ● ● trimming of formyl group from formyl-Met proteolytic removal of N-terminal Met by aminopeptidases acetylation lipidation (myristoylation) http://bass.bio.uci.edu/~hudel/bs99a/lecture26/lecture7_2.html (2 of 4)5/24/2007 12:55:35 PM Covalent Modifications (7.2) Modifications involving the carboxy terminus: ● ● amidation of C-terminal glycine attachment of membrane anchors http://bass.bio.uci.edu/~hudel/bs99a/lecture26/lecture7_2.html (3 of 4)5/24/2007 12:55:35 PM Covalent Modifications (7.2) Next: Side Chain Modifications Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture26/lecture7_2.html (4 of 4)5/24/2007 12:55:35 PM Side Chain Modifications (7.3) Side Chain Modifications Modifications involving amino acid side chains: ● disulfide cross-linking ● lysinonorleucine cross-linking (collagen) http://bass.bio.uci.edu/~hudel/bs99a/lecture26/lecture7_3.html (1 of 13)5/24/2007 12:55:45 PM Side Chain Modifications (7.3) ● Phosphorylation of hydroxyls by kinases (serine, threonine, tyrosine): http://bass.bio.uci.edu/~hudel/bs99a/lecture26/lecture7_3.html (2 of 13)5/24/2007 12:55:46 PM Side Chain Modifications (7.3) http://bass.bio.uci.edu/~hudel/bs99a/lecture26/lecture7_3.html (3 of 13)5/24/2007 12:55:46 PM Side Chain Modifications (7.3) Glycogen phosphorylase is the ultimate enzyme in a cascade which catalyzes the degradation of glycogen to glucose-1-phosphate. Phosphorylation of phosphorylase occurs on serine-14 and converts the inactive phosphorylase b to the active phosphorylase a. http://bass.bio.uci.edu/~hudel/bs99a/lecture26/lecture7_3.html (4 of 13)5/24/2007 12:55:46 PM Side Chain Modifications (7.3) Phosphorylation is reversible and is used in many pathways to control activity. Enzymes that add a phosphate to a hydroxyl side chain are commonly called kinases. Enzymes that remove a phosphate from a phosphorylated side chain are called phosphatases. Glycosylation ● There are two basic types of glycosylation which occur on: asparagines (N-linked, see (a) below) and serines and threonines (O-linked, see (b) below) ❍ ❍ http://bass.bio.uci.edu/~hudel/bs99a/lecture26/lecture7_3.html (5 of 13)5/24/2007 12:55:46 PM Side Chain Modifications (7.3) (b) N-Acetylgalactosamine ● ● Covalently attached to the polypeptide as oligosaccharide chains containing 4 to 15 sugars Sugars frequently comprise 50% or more of the total molecular weight of a glycoprotein http://bass.bio.uci.edu/~hudel/bs99a/lecture26/lecture7_3.html (6 of 13)5/24/2007 12:55:46 PM Side Chain Modifications (7.3) ● ● ● ● Most glycosylated proteins are either secreted or remain membrane-bound Glycosylation is the most abundant form of post-translational modification Glycosylation confers resistance to protease digestion by steric protection Important in cell-cell recognition N-linked glycosylation on asparagine (Asn) side chains: ● ● ● ● ● ● an alkali-stable bond between the amide nitrogen of asparagine and the C-1 of an amino sugar residue occurs co-translationally in the endoplasmic reticulum (ER) during synthesis lipid-linked oligosaccharide complex is transferred to polypeptide by oligosaccharyl transferase target sequence or consensus site on protein is Asn-X-Ser/Thr further processing in Golgi apparatus Examples: ❍ Heavy chain of immunoglobulin G (IgG) ❍ Hen ovalbumin ❍ Ribonuclease B O-linked glycosylation on serine (Ser) or threonine (Thr) side chains ● ● ● ● an alkali-labile bond between the hydroxyl group of serine or threonine and an amino sugar carried out by a class of membrane-bound enzymes called glycosyl transferases which reside in the endoplasmic reticulum (ER) or the Golgi apparatus nucleotide-linked monosaccharides added to protein side chain one at a time Example: Blood group antigens on erythrocyte surface: ❍ The A antigen and B antigen are pentasaccharides which differ in composition of the 5th sugar residue th residue and does not ❍ The O substance is a tetrasaccharide which is missing the 5 elecit an antibody response (non-antigenic). http://bass.bio.uci.edu/~hudel/bs99a/lecture26/lecture7_3.html (7 of 13)5/24/2007 12:55:46 PM Side Chain Modifications (7.3) , http://bass.bio.uci.edu/~hudel/bs99a/lecture26/lecture7_3.html (8 of 13)5/24/2007 12:55:46 PM Side Chain Modifications (7.3) Blood Type Glycolsyl transferase Antibodies Against (Antigen) Can Safely Receive Blood from Can Safely Donate Blood to O neither A&B O O, A, B & AB A UDP-GalNAc B O&A A & AB B UDP-Gal A O&B B & AB AB both neither O, A, B, & AB AB Examples of monosaccharides used in glycosylation ● Prosthetic group attachment (heme, retinal etc.) http://bass.bio.uci.edu/~hudel/bs99a/lecture26/lecture7_3.html (9 of 13)5/24/2007 12:55:46 PM Side Chain Modifications (7.3) Heme group, attached to histidine side chain via Fe. http://bass.bio.uci.edu/~hudel/bs99a/lecture26/lecture7_3.html (10 of 13)5/24/2007 12:55:46 PM Side Chain Modifications (7.3) Retinal attached to lysine side chain via covalent Schiff base linkage. http://bass.bio.uci.edu/~hudel/bs99a/lecture26/lecture7_3.html (11 of 13)5/24/2007 12:55:46 PM Side Chain Modifications (7.3) Processing of pre-pro-insulin to active insulin http://bass.bio.uci.edu/~hudel/bs99a/lecture26/lecture7_3.html (12 of 13)5/24/2007 12:55:46 PM Side Chain Modifications (7.3) ● ● ● ● Pre-pro-insulin is synthesized as a random coil on membrane-associated ribosomes After membrane-transport the leader sequence (yellow) is cleaved off by a protease and the resulting pro-insulin folds into a stable conformation. Disulfide bonds form between cysteine side chains. The connecting sequence (red) is cleaved off to form the mature and active insulin molecule. Next: Noncovalent Modifications Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture26/lecture7_3.html (13 of 13)5/24/2007 12:55:46 PM Noncovalent Modifications (7.4) Noncovalent Modifications Addition of metal ions and co-factors: Nearly 50% of all proteins contain metal ions Metal ions play regulatory as well as structural roles ● ● ● ● Calcium (Ca++): very important intra-cellular messenger, i.e. calmodulin Magnesium (Mg++): ATP enzymes Copper (Cu++), Nickel (Ni+), Iron (Fe++) Zinc (Zn++): Zinc finger domains are used for DNA recognition: http://bass.bio.uci.edu/~hudel/bs99a/lecture26/lecture7_4.html (1 of 3)5/24/2007 12:55:58 PM Noncovalent Modifications (7.4) A zinc finger domain: Zn++ is bound by two cysteine and two histidine residues. Zinc finger domains interact in the major groove with three consecutive bases from one strand of duplex Bform DNA. Modifications involving tertiary structure (protein fold) Enzymes called molecular chaperones are responsible for detecting mis-folded proteins. Chaperones only bind mis-folded proteins that exhibit large hydrophobic patches on their surfaces. http://bass.bio.uci.edu/~hudel/bs99a/lecture26/lecture7_4.html (2 of 3)5/24/2007 12:55:58 PM Noncovalent Modifications (7.4) Subunit multimerization Many enzymes are only functional as multimeric units, either as homo- or hetero-oligomers. Example: ribosomes! Next: Chaperone-Assisted Protein Folding Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture26/lecture7_4.html (3 of 3)5/24/2007 12:55:58 PM Chaperone-Assisted Protein Folding (7.5) Chaperone-Assisted Protein Folding Quote: "The traditional role of the human chaperone described in biochemical terms is to prevent improper interactions between potentially complementary surfaces, and to disrupt any improper liaisons which occur." Chaperones: ● ● ● ● ● ● ● ● ● Mediate folding and assembly. Do not convey steric information. Do not form part of the final structure. Suppress non-productive interactions by binding to transiently exposed portions of the polypeptide chain. First identified as heat shock proteins (Hsp). Hsp expression is elevated when cells are grown at higher-than-normal temperatures. Stabilize proteins during synthesis. Assist in protein folding by binding and releasing unfolded/mis-folded proteins. Use an ATP-dependent mechanism. Major types of chaperones: ● Hsp70 (cytoplasm, ER, chloroplasts, mitochondria): ❍ thought to bind and stabilize the nascent polypeptide chain as it is being extruded from the ribosome. ❍ also involved in "pulling" newly synthesized polypeptide into ER lumen. http://bass.bio.uci.edu/~hudel/bs99a/lecture26/lecture7_5.html (1 of 3)5/24/2007 12:56:05 PM Chaperone-Assisted Protein Folding (7.5) ● Hsp60 (mitochondria, chloroplasts): ❍ forms large 28-subunit complexes called GroEL http://bass.bio.uci.edu/~hudel/bs99a/lecture26/lecture7_5.html (2 of 3)5/24/2007 12:56:05 PM Chaperone-Assisted Protein Folding (7.5) Next: Selenocysteine Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture26/lecture7_5.html (3 of 3)5/24/2007 12:56:05 PM Selenocysteine (7.6) Selenocysteine: the 21st Amino Acid? Although the element selenium was discovered in 1817 by Berzelius it was only shown over 100 years later to be an essential micronutrient in all three lines of decent. Subsequent analysis of several enzymes that catalyze oxidation-reduction reactions showed that selenium occurs in the form of the unusual amino acid selenocysteine. How this amino acid is incorporated into the protein was unclear in these days. Today it is well established that the incorporation of selenocysteine is co-translational. Interestingly, the base triplet encoding this amino acid is UGA, a codon that normally functions as a STOP signal in translation. Since this codon has still retained its "normal" function in all organisms known to synthesize selenocysteine-containing proteins, the incorporation of this amino acid requires a specific pathway. This http://bass.bio.uci.edu/~hudel/bs99a/lecture26/lecture7_6.html (1 of 5)5/24/2007 12:56:11 PM Selenocysteine (7.6) pathway has been elucidated for the bacterium E. coli by August Böck and coworkers. General pathway There are one cis-acting element and four trans-acting factors involved in this incorporation. The cis-acting element is a specific stem-loop structure of the mRNA directly 3' of the UGA codon. The four trans-acting factors are: ● ● ● ● a specific tRNASec which is charged with serine by serinyl-tRNA synthetase the enzyme selenophosphate synthetase which generates inorganic selenophosphate the enzyme selenocysteine synthetase which uses selenophosphate to convert seryl-tRNASec to selenocysteyl-tRNASec a specific translation factor (SelB) that substitutes for EF-Tu and recognizes the cis-acting element In the first step the specific tRNASec is charged by the normal seryltRNA synthetase with serine, and that serine is subsequently converted to selenocysteine by the enzyme selenocysteine synthetase. The low molecular weight selenium donor - selenophosphate - is provided by the action of the selenophosphate synthetase. Finally, the selenocysteyl-tRNASec is recognized by a specific translation factor http://bass.bio.uci.edu/~hudel/bs99a/lecture26/lecture7_6.html (2 of 5)5/24/2007 12:56:11 PM Selenocysteine (7.6) SelB that delivers it to a UGA codon at the ribosomal A-site only in the presence of the stem-loop structure downstream of the codon on the mRNA. tRNASec tRNASec differs in several aspects from the consensus for canonical tRNAs. In particular it includes an acceptor stem elongated by extra one base pair, an elongated D-arm with only four nucleotides in the loop, and a UCA anticodon. These differences ensure that tRNASec is not recognized by the normal translation factor EF-Tu thus preventing mis-incorporation. http://bass.bio.uci.edu/~hudel/bs99a/lecture26/lecture7_6.html (3 of 5)5/24/2007 12:56:11 PM Selenocysteine (7.6) The two structures shown here are (a) tRNASec and (b) tRNASer of E. coli. Residues which are normally conserved in tRNA are shown circled. Identity nucleotides in tRNASer and their counterparts in tRNASec are shown boxed. tRNASec is the longest tRNA species known consisting of 95 nucleotides. Selenophosphate synthetase This enzyme catalyses the synthesis of the low molecular weight donor selenophosphate using ATP as the phosphate donor. Interestingly, the gamma-phosphate is transferred to selenium whereas the beta-phosphate is released, leaving AMP. Selenocysteine synthetase This enzyme possesses a prosthetic group (pyridoxal phosphate). The active enzyme is composed of ten identical subunits arranged as a stack of two five-membered rings. It converts the serine attached to tRNASec to selenocysteine. Translation factor SelB This translation factor substitutes for elongation factor EF-Tu in the specific incorporation of selenocysteine. This protein is considerable longer than its regular counterpart. The N-terminal half of the protein resembles EF-Tu in structure and function - GTP and tRNA binding whereas the C-terminal half is responsible for the recognition of the http://bass.bio.uci.edu/~hudel/bs99a/lecture26/lecture7_6.html (4 of 5)5/24/2007 12:56:11 PM Selenocysteine (7.6) specific stem-loop structure adjacent to the UGA codon. A complex between SelB, GTP, selenocysteyl-tRNASec and the stem-loop structure of the mRNA has been detected and shown to be functionally necessary. Next: Summary Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture26/lecture7_6.html (5 of 5)5/24/2007 12:56:11 PM Summary (7.7) Summary ● ● ● Post-translational processing controls folding, targeting, activation and stability of proteins Co- and post-translational modifications increase diversity and functionality of proteins Common forms of co- or post-translational modifications are: proteolysis phosphorylation glycosylation metal binding ❍ ❍ ❍ ❍ ● ● Chaperones mediate folding and assembly of newly synthesized proteins Selenocysteine is sometimes called the 21st amino acid uses one of the three STOP codons has its own tRNASec requires other factors, including a cis-acting element on the mRNA ❍ ❍ ❍ Next Lecture: Protein Trafficking/Targeting Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture26/lecture7_7.html5/24/2007 12:56:16 PM Protein Trafficking/Targeting (8.1) Lecture 8 Protein Trafficking/Targeting Protein targeting is necessary for proteins that are destined to work outside the cytoplasm. Protein targeting is more complex in eukaryotes because of the presence of many intracellular compartments. Prokaryotic protein targeting (secretion) http://bass.bio.uci.edu/~hudel/bs99a/lecture27/index.html (1 of 3)5/24/2007 12:56:22 PM Protein Trafficking/Targeting (8.1) The chaperone protein SecB binds to the nascent polypeptide chain to prevent premature folding which would make transport across the plasma membrane impossible. SecE and SecY are transmembrane components which form a pore in the membrane through which the still unfolded polypeptide is threaded. The translocation process is energy-dependent (ATP) and is driven by SecA. Once the protein has passed through the pore, the signal sequence is cleaved off by an extracellular, membrane-bound protease. N-terminal signal sequences of representative secreted prokaryotic proteins. Protein -20 http://bass.bio.uci.edu/~hudel/bs99a/lecture27/index.html (2 of 3)5/24/2007 12:56:22 PM -10 -1 +1 Protein Trafficking/Targeting (8.1) Leucinebinding protein MKANAKTIIAGMIALAISHTAMA EE... Pre-alkaline phosphatase MKQSTIALALLPLLFTPVTKA RT... Prelipoprotein MKATKLVLGAVILGSTLLAG CS... Hydrophobic residues in red. Next: Eukaryotic Protein Targeting Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture27/index.html (3 of 3)5/24/2007 12:56:22 PM Eukaryotic Protein Targeting (8.2) Eukaryotic Protein Targeting Targeting in eukaryotes is necessarily more complex due to the multitude of internal compartments: ● ● ● ● ● ● ● ● nucleus mitochondria peroxisomes chloroplasts endoplasmic reticulum (ER) Golgi lysosomes secretory granules The signals involved are also called sorting signals. They are regions on the targeted protein with certain amino acid sequences. These signals interact with specific receptors, either on the target organelle or a carrier protein. http://bass.bio.uci.edu/~hudel/bs99a/lecture27/lecture8_2.html (1 of 4)5/24/2007 12:56:27 PM Eukaryotic Protein Targeting (8.2) There are two basic forms of targeting pathways: ● ● post-translational targeting: ❍ nucleus ❍ mitochondria ❍ chloroplasts ❍ peroxisomes co-translational targeting (secretory pathway): ❍ ER ❍ Golgi ❍ lysosomes ❍ plasma membrane ❍ secreted proteins http://bass.bio.uci.edu/~hudel/bs99a/lecture27/lecture8_2.html (2 of 4)5/24/2007 12:56:27 PM Eukaryotic Protein Targeting (8.2) In the absence of targeting signals, a protein will remain in the cytoplasm: ● ● ● ● translational machinery metabolic enzymes cytoskeletal proteins many signal transduction proteins Nuclear targeting: ● Unusual since 2-way traffic: ❍ in: proteins, DNA ■ DNA & RNA polymerases ■ transcriptions factors ■ histones etc. ❍ out: mRNA, tRNA, rRNA http://bass.bio.uci.edu/~hudel/bs99a/lecture27/lecture8_2.html (3 of 4)5/24/2007 12:56:27 PM Eukaryotic Protein Targeting (8.2) ● Proteins are not transported through the nuclear membrane but rather through a complex pore called the nuclear pore: ❍ comprised of about 100 different proteins ❍ proteins smaller than 20 kDa move by diffusion ❍ proteins larger than 20 kDa move by selective transport (nuclear localization signal) ■ cluster of 4-8 positively charged amino acids (example: PKKKRLV) ■ signal sequence binds to receptor on the pore called importin Mitochondrial targeting: ● ● not well understood usually by post-translational targeting Lysosomal targeting: ● ● ● ● Lysosomes are organelles that store enzymes which rapidly degrade other proteins and nucleic acids. A famous target sequence is "KDEL" Initial targeting via secretory pathway Final targeting occurs in the Golgi Next: The Secretory Pathway Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture27/lecture8_2.html (4 of 4)5/24/2007 12:56:27 PM The Golgi (8.2b) The secretory pathway ER targeting (secretory pathway) ● co-translational insertion of protein into or through ER membrane via attached ribosomes (rough ER): ❍ signal sequence of 16-30 amino acids at N-terminus (hydrophobic) ❍ emerging signal sequence of nascent protein on free ribosome binds to signal recognition particle (SRP) -- translation is arrested. ■ SRPs consist of 6 proteins and one RNA molecule (7S RNA). ■ The SRP-signal sequence-mRNA-ribosome complex docks with receptor on ER membrane. ❍ signal sequence crosses ER membrane. ❍ translation continues with polypeptide chain being pulled into the ER lumen. http://bass.bio.uci.edu/~hudel/bs99a/lecture27/lecture8_2b.html (1 of 5)5/24/2007 12:56:56 PM The Golgi (8.2b) While in the ER, many proteins undergo the first stages of glycosylation. Most proteins then migrate inside vesicles from the ER and enter the cis face of the Golgi where further processing and final sorting occurs: The Golgi Complex http://bass.bio.uci.edu/~hudel/bs99a/lecture27/lecture8_2b.html (2 of 5)5/24/2007 12:56:56 PM The Golgi (8.2b) The Golgi is responsible for further processing and final sorting of proteins. One example is the formation of primary and secondary lysosomes: ● Primary lysosomes bud from the trans face of the Golgi and subsequently ❍ undergo exocytosis (A) ❍ fuse with vesicles to digest their contents (B & C) ❍ rupture, causing autolysis (D) Overview of Trafficking http://bass.bio.uci.edu/~hudel/bs99a/lecture27/lecture8_2b.html (3 of 5)5/24/2007 12:56:56 PM The Golgi (8.2b) http://bass.bio.uci.edu/~hudel/bs99a/lecture27/lecture8_2b.html (4 of 5)5/24/2007 12:56:56 PM The Golgi (8.2b) Next: Targeted Protein Degradation Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture27/lecture8_2b.html (5 of 5)5/24/2007 12:56:56 PM Targeted Protein Degradation (8.3) Targeted Protein Degradation Why are proteins degraded? In order to keep a cell working it needs to remove: ● ● ● ● incorrectly synthesized proteins (with errors in amino acid sequence) damaged proteins (i.e. oxidative damage) cell-cycle specific proteins other signaling proteins which are no longer necessary One mechanism of protein degradation is via lysosomes. Lysosomes are acidic vesicles that contain about 50 different enzymes involved in degradation: ● ● ● ● ● ● ● ● proteases (cathepsins): cleave peptide bonds phosphatases: remove covalently bound phosphates nucleases: cleave DNA/RNA lipases: cleave lipid molecules carbohydrate-cleaving enzymes: remove covalently bound sugars from glycoproteins Lysosomes often secrete their contents into the extracellular medium via exocytosis. Lysosomes can also target damaged organelles in a process called autophagy. Sometimes, lysosomes are triggered to rupture inside a cell, resulting in autolysis, http://bass.bio.uci.edu/~hudel/bs99a/lecture27/lecture8_3.html (1 of 3)5/24/2007 12:57:17 PM Targeted Protein Degradation (8.3) also called apoptosis or programmed cell death. Another major mechanism is via ubiquitin labeling of surplus proteins: ● ● ● ● Ubiquitin (a small 76-residue protein) is attached to the protein: ❍ First, an activating enzyme attaches itself to the carboxy terminus of free ubiquitin in an ATP-dependent process. ❍ Then, the activated ubiquitin is transferred onto a second enzyme which at the same time recognizes damaged proteins. ❍ The activated ubiquitin is then covalently linked to lysine residues on the surface of the damaged protein. These ubiquitin-tagged proteins are now recognized by specific proteases in the cytosol which in turn cleave and degrade the tagged protein. These proteases are combined in a very large protein complex called the proteasome. The proteasome (20S) is comprised of 28 subunits and has a molecular weight of 700 kDa: http://bass.bio.uci.edu/~hudel/bs99a/lecture27/lecture8_3.html (2 of 3)5/24/2007 12:57:17 PM Targeted Protein Degradation (8.3) Next: Summary Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture27/lecture8_3.html (3 of 3)5/24/2007 12:57:17 PM Summary (8.4) Summary ● ● ● Targeting of newly synthesized proteins is an integral component of protein synthesis. In prokaryotes, targeting is usually achieved by an N-terminal signal sequence of about 20 mostly hydrophobic amino acids. In eukaryotes, targeting is more complex due to the large number of different cellular compartments: Nuclear targeting via the nuclear pore using a nuclear localization signal. ER targeting (secretory pathway) via N-terminal signal sequences using SRPs with subsequent attachment to the ER, Followed by transport to the Golgi complex ❍ ❍ ❍ ● Protein degradation of damaged or obsolete proteins is carried out by lysosomes, vesicles filled with degradating enzymes in a ubiqutin-dependent process by specific proteases in a large cytosolic complex called the proteasome. ❍ ❍ Last Lecture: Expression Systems for Recombinant Proteins Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture27/lecture8_4.html5/24/2007 12:57:33 PM Final Exam (9.0) Final Exam, BS99A Final Exam, BS99B ● ● ● ● ● ● ● There will be assigned seating (charts posted at PSLH doors at 10:00 am). Bring your student ID. Bring a pencil. No calculators, no pagers, no hats, etc... Multiple choice AND essay questions will cover lectures since the mid-term. After 12:15pm, please don't get up until all exams have been collected. Questions regarding grading should be directed in writing and with a detailed explanation to the TA. Please fill out teaching evaluations (for my section only) and turn them in after today's lecture. Last Lecture: Expression Systems for Recombinant Proteins http://bass.bio.uci.edu/~hudel/bs99a/lecture28/index.html (1 of 2)5/24/2007 12:57:37 PM Final Exam (9.0) Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture28/index.html (2 of 2)5/24/2007 12:57:37 PM Expression Systems for Recombinant Proteins (9.1) Lecture 9 Expression Systems for Recombinant Proteins Recombinant protein expression is the foundation of today's biomolecular research and the thriving Biotech industry. Goal: Overproduction of proteins for structural & functional studies and for medical & industrial applications. As a 199 research student chances are high that you will engage in some aspect of recombinant protein expression. http://bass.bio.uci.edu/~hudel/bs99a/lecture28/lecture9_1.html (1 of 4)5/24/2007 12:57:49 PM Expression Systems for Recombinant Proteins (9.1) These techniques rely on: ● the universality of the Genetic Code ● knowing the Genetic Code ● the relative similarity of the translational machinery (ribosome) ● the rapid progress in molecular biology/genetic engineering over the past few decades: ❍ sequence-specific nucleic acid hybridization (1961) ❍ sequence-specific cleavage of DNA (1962) ❍ DNA cloning/amplification (early 70s) ❍ DNA sequencing (mid 70s) ❍ Cutting and pasting pieces of DNA from one source into another: ■ excise with sequence-specific restriction endonuclease (more than 100 known, normally employed for defense by bacteria) ■ hybridize sticky ends ■ reform covalent phosphodiester bonds with DNA ligase (1967) http://bass.bio.uci.edu/~hudel/bs99a/lecture28/lecture9_1.html (2 of 4)5/24/2007 12:57:49 PM Expression Systems for Recombinant Proteins (9.1) Examples of Recombinant Protein Products ● ● ● Hormones ❍ Insulin: Diabetes ❍ Human thyroid stimulating hormone Blood clotting factors ❍ Coagulation factor VIII : hemophilia A. ❍ Coagulation factor IX: hemophilia B. Interferons ❍ interferon-(alpha)-2a: chronic hepatitis C. ❍ gamma interferon: hepatitis B, C, herpes and viral enteritis. http://bass.bio.uci.edu/~hudel/bs99a/lecture28/lecture9_1.html (3 of 4)5/24/2007 12:57:49 PM Expression Systems for Recombinant Proteins (9.1) ● ● Immunization agents ❍ Hepatitis B vaccine: a non-infectious vaccine derived from Hepatitis B surface antigen (HBSA) produced in yeast cells. Research enzymes ❍ Restriction endonucleases ❍ Endoglycosidases: PNGase F Next: DNA Cloning Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture28/lecture9_1.html (4 of 4)5/24/2007 12:57:49 PM DNA Cloning (9.2) DNA Cloning Cloning: 1. 2. 3. 4. 5. 6. 7. 8. 9. 10. Cell fractionation by conventional chromatographic methods to yield about a microgram of protein. The protein is analyzed to yield the identity of the first 30 amino acids - its N-terminal amino acid sequence. The Genetic Code is used to predict nucleotide sequences corresponding to this amino acid sequence. DNA fragments complementary to these sequences (oligomers of 1520 bases) are chemically synthesized. The DNA fragments are then hybridized (base-paired) with total cellular mRNA. Long cDNA segments are produced from mRNAs complementary to the DNA fragments using the enzyme reverse transcriptase. Large amounts of this cDNA is obtained by cloning into plasmids (amplification). Selection of the right clone (several steps). Finally, the cloned cDNA is incorporated into an expression vector or plasmid and transferred into bacterial or yeast cells. This is the starting point for scaled-up production of large amounts of the protein. http://bass.bio.uci.edu/~hudel/bs99a/lecture28/lecture9_2.html (1 of 6)5/24/2007 12:57:54 PM DNA Cloning (9.2) Expression vectors or plasmids must contain: ● ● ● ● origin of replication: DNA polymerase selectable marker(s): antibiotic resistance promoter: recognized by RNA polymerase multiple clonig sites (restriction enzyme sites): cutting/pasting of DNA fragments Plasmids are small circular molecules of extrachromosomal, double-stranded DNA. They occur naturally in both bacteria and yeast where they replicate as independent units. Unlike chromosomal DNA, plasmids usually occur as multiple copies. http://bass.bio.uci.edu/~hudel/bs99a/lecture28/lecture9_2.html (2 of 6)5/24/2007 12:57:54 PM DNA Cloning (9.2) Restriction endonuclease EcoRI cuts double-stranded DNA and generates sticky ends. Sticky ends hybridize (base-pair) to each other and DNA ligase reforms covalent phosphodiester backbone. http://bass.bio.uci.edu/~hudel/bs99a/lecture28/lecture9_2.html (3 of 6)5/24/2007 12:57:54 PM DNA Cloning (9.2) A piece of DNA can be inserted into a plasmid if both the circular plasmid and the source of DNA have recognition sites for the same restriction endonuclease. http://bass.bio.uci.edu/~hudel/bs99a/lecture28/lecture9_2.html (4 of 6)5/24/2007 12:57:54 PM DNA Cloning (9.2) General scheme for insertion of gene of interest into an expression plasmid. http://bass.bio.uci.edu/~hudel/bs99a/lecture28/lecture9_2.html (5 of 6)5/24/2007 12:57:54 PM DNA Cloning (9.2) Next: Expression Systems Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture28/lecture9_2.html (6 of 6)5/24/2007 12:57:54 PM Expression Systems (9.3) Expression Systems The DNA on the plasmid is transcribed to mRNA which in turn is translated to protein. Types of expression systems: ● ● ● ● ● Bacterial: plasmids, phages Yeast: expression vectors: plasmids, yeast artifical chromosomes (YACs) Insect cells: baculovirus, plasmids Frog oocytes: injected mRNA Mammalian: ❍ viral expression vectors (gene therapy): ■ SV40 ■ vaccinia virus ■ adenovirus ■ retrovirus ❍ Stable cell lines (CHO, HEK293) http://bass.bio.uci.edu/~hudel/bs99a/lecture28/lecture9_3.html (1 of 3)5/24/2007 12:57:59 PM Expression Systems (9.3) Bacterial expression systems: ● Usually E. coli ● A specific gene on a plasmid can produce 1-30% of the total protein. ● ● ● For native protein: ❍ Ligate gene to bacterial promoter on plasmid. ❍ Watch for frame shift! For fusion protein: ❍ DNA for gene of interest is inserted after the 3' or before the 5' terminus of "carrier" gene (GST, GFP). ❍ Watch for frame shift! ❍ Advantages over native protein expression: ■ synthesized at high levels like a normal bacterial gene ■ often results in more stable product than native protein ■ fusion protein is generally larger than most E. coli proteins: easy identification & purification ■ exploit functional features of carrier protein in purification ■ Caveat: often low yields when cleaving carrier from target protein with protease For His-tagged protein: ❍ By mutation introduce multiple histidine codons (6 or more) at the N-terminus or C-terminus ❍ Purification over Nickel column: only proteins with poly-His tag will bind tightly http://bass.bio.uci.edu/~hudel/bs99a/lecture28/lecture9_3.html (2 of 3)5/24/2007 12:57:59 PM Expression Systems (9.3) Common problems with bacterial expression systems: ● ● ● ● ● Low expression levels: ❍ change promoter ❍ change plasmid ❍ change cell type ❍ add rare tRNAs for rare codons on second plasmid Severe protein degradation: ❍ use proteasome inhibitors and other protease inhibitors ❍ try induction at lower temperature Missing post-translational modification: co-express with kinases etc. Glycosylation will not be carried out: ❍ use yeast or mammalian expression system Misfolded protein (inclusion bodies): ❍ co-express with GroEL, a chaperone ❍ try refolding buffers Next: Summary Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture28/lecture9_3.html (3 of 3)5/24/2007 12:57:59 PM Summary (9.4) Summary ● ● ● ● Recombinant protein expression relies on the universality of the Genetic Code It employs a host of sophisticated techniques to analyze, manipulate and copy DNA (genetic engineering) The gene of interest (DNA) is cloned into an expression plasmid or vector The DNA on the plasmid is transcribed to mRNA which in turn is translated to protein Next: Final Exam Please report typos, errors etc. by EMAIL (mention the title of this page). http://bass.bio.uci.edu/~hudel/bs99a/lecture28/lecture9_4.html5/24/2007 12:58:05 PM http://bass.bio.uci.edu/~hudel/bs99a/final.html Final Exam for BS99A Final Exam Key for Dr. Osborne's section Final Exam Key for the section on Protein Synthesis Final Grades The final exam will only cover lectures 11 to 28. It will contribute up to 60 out of a total of 100 points towards the final grade. The only valid excuse for missing this exam is a substantiated medical emergency or written arrangements made with Dr. Luecke. Requests and reasons for missing the exam must be submitted in writing. Makeup exams will be oral and, unlike the written exam, comprehensive! Questions regarding grading need to be directed before noon on June 30 in writing and with a detailed explanation to one of the TAs. Only if no satisfactory solution can be found with the TAs, a written request (email or paper) with a detailed explanation regarding the circumstances should be sent to Dr. Luecke. Dr. Hartmut Luecke 3205 McGaugh Hall ZOT 3900 http://bass.bio.uci.edu/~hudel/bs99a/final.html5/24/2007 12:58:08 PM Bio 99A 2005 Key Osborne Answers: Version1/Version 2 1/14 ). (1 point) Complete the following sentence with 3-5 words The alpha (α) subunit of E.coli RNA polymerase functions in transcription by … binding upstream DNA non-sepcifically and binding to other regulatory proteins (like CAP)______ + 2/15). (1 point) In the regulation for the Lac operon of E. coli, the wild type I is recessive to what two important types of lac operon regulatory mutations (use no more than two words ) IS and Oc 3/16 ). (total of 3 points, one-two sentences of no more than 25-30 total words, no partial credit). In the TRP operon of E. coli, if there was an insertion mutation in the leader DNA that resulted in an additional 40 random nucleotides inserted into the leader RNA sequence between stem region 1 and stem region 2, how would attenuation likely be affected when intracellular Tryptophan levels are high (would attenuation be increased or decreased-1 point) and why ? (2 points)) attenuation would decrease, The insertion means that when the ribosome does not stall, it cannot cover region 2 effectively so it is free to more frequently pair with region 3. 4/17 ). (1 point, one to three words) RNA polymerase III transcribes which major class of RNA in eukaryotes? tRNA 5/18 ). (1 point-no partial credit) When lactose is added to a bacterial culture of E. coli with each of the following genotypes, what would be the pattern for β-galactosidase expression for each haploid strain below (a and b below) note that the solid line on the answer sheet provides the pattern for βgalactosidase enzyme activity in a strain that is completely wild type (I+ P+O+Z+Y+) - a). I P+O+Z+Y+ - b). IS P+OcZ Y+ Copyright © 2005 Unless otherwise indicated, all materials on these pages are copyrighted by Drs.Timothy F Osborne and Hartmut Luecke. All rights reserved. No part of these pages, either text or image may be used for any purpose other than personal use. Therefore, reproduction, modification, storage in a retrieval system or retransmission, in any form or by any means, electronic, mechanical or otherwise, for reasons other than personal use, is strictly prohibited without prior written permission. Bio 99A 2005 Key Osborne 6/19). (1 point, one word) When the two above strains are combined to form a partial diploid strain - containing both Lac operons, what is the phenotype (Lac + or Lac ) - Lac 7/20). (2 points, no partial credit) If the TRP regulatory gene (TRP R) encoded a protein that stimulated RNA polymerase binding to the promoter (positive control mechanism) and yet the operon was still repressed by the accumulation of tryptophan what would be the most likely effect of tryptophan on the DNA binding properties of the TRP R regulatory protein? TRP would decrease TRPR binding to operator 8/21 ). (one point for each-total of 3 points) As the mechanism of attenuation was described in class, there are three molecular features that are absolutely required. List them 1).__Coupled transcription (RNA synthesis) and translation (protein synthesis_ 2).__Alternative secondary structures in leader mRNA___ 3).__Clustered (2 or more) codons for the regulatory amino acid in the leader mRNA_ 9/22 ). (2 points-no partial credit) In the binding of the Lac I protein to the DNA, we discussed a general equlibrium based method for evaluating proteins binding to DNA. These same principles apply to RNA polymerase (RNAP) from E. coli binding to different promoters. If the solid line on the graph provided on the answer sheet describes the binding of RNA polymerase to a promoter that is identical to the “consensus sequence”, insert a graph that would describe RNA polymerase binding to a weak promoter such as from the Lac operon –use a dashed line graph? 10/21). (1 point) The core E. coli RNA polymerase contains which subunits and how many copies of each? 2-alpha, 1-beta, 1-beta prime Copyright © 2005 Unless otherwise indicated, all materials on these pages are copyrighted by Drs.Timothy F Osborne and Hartmut Luecke. All rights reserved. No part of these pages, either text or image may be used for any purpose other than personal use. Therefore, reproduction, modification, storage in a retrieval system or retransmission, in any form or by any means, electronic, mechanical or otherwise, for reasons other than personal use, is strictly prohibited without prior written permission. Bio 99A 2005 Key Osborne 11/22 ). (1 point) What is the fundamental difference between RNA polymerase and DNA polymerase (besides one being involved in DNA replication and one being involved in transcription). ? One sentence of up to 15-16 words and be sure to refer to each polymerase in your answer. (1 point) _RNAP can initiate de nove0. DNAP require a primer with 3’OH for extension 12/23 ). (1 point) In protein assembly onto a core eukaryotic promoter for RNA polymerase II (on to the TATA box) which proteins are bound at the promoter before RNA polymerase is recruited? TF IID, A and B (note TBP is only part of TFIID) 13/24 ). Consider a strain of E. coli with the following genotype for the Lac operon (I- P+OCZ+Y+) A). (1 point) use a dashed line graph to draw the pattern of β-galactosidase expression over time when only Lactose is added (the solid line describes what would happen to the wild type strain (I+ P+O+Z+Y+) B). (1 point) use a dashed line graph to draw the pattern for β-galactosidase expression over time when lactose and a high concentration of glucose are added together (the solid line describes what would happen to the wild type strain when only lactose is added (I+ P+O+Z+Y+) C). (3 points) Based on what we discussed in class, explain the difference between A and B above (one to two sentences total of 25 words) C). at high glucose, cAMP levels are low and CAP does not bind to its DNA site and the weak lac promoter is not expressed 14/25). (1 point, up to three words) Which specific eukaryotic RNA polymerase is inhibited only when very high concentrations of the drug alpha (α) amanitin are added to a transcription reaction? RNA Polymerase I Copyright © 2005 Unless otherwise indicated, all materials on these pages are copyrighted by Drs.Timothy F Osborne and Hartmut Luecke. All rights reserved. No part of these pages, either text or image may be used for any purpose other than personal use. Therefore, reproduction, modification, storage in a retrieval system or retransmission, in any form or by any means, electronic, mechanical or otherwise, for reasons other than personal use, is strictly prohibited without prior written permission. Bio 99A 2005 Key Osborne 15/26 ). (1 point, one word) In mammalian cells, the type of chromatin associated actively transcribed genes is called euchromatin 16/27 ). (1 point, one sentence, 10-12 words) What is the role of TF IIA in transcription? Stabilize TBP bound to TATA box (not specific enough to say just stabilize TBP) 17/28 ). (1 point, one to two words) If DNA was wrapped around each nucleosome three times, how many total basepairs of DNA would be associated with each nucleosome? 18/31). (3 points, two sentences, 20-30 words) On the planet of Tatuine, the Empire Research Corporation has found a new life force that threatens the stability of the Empire. When researchers evaluate RNA expression from this new organism they perform a RNA synthesis experiment very similar to what we described in Bio 99. When RNA was synthesized in the absence or presence of different concentrations of the inhibitor alpha (α) amanitin the results were analyzed by gel electrophoresis as displayed in the figure below. What can you conclude about the RNA synthesizing enzymes of this new life force? Two a-amanitin sensitive enzymes, one transcribes tRNAs and rRNAs and is inhibited by low drug concentration and a second enzyme that transcribes mRNAs and is inhibited by only a high alpha-amanitin level Copyright © 2005 Unless otherwise indicated, all materials on these pages are copyrighted by Drs.Timothy F Osborne and Hartmut Luecke. All rights reserved. No part of these pages, either text or image may be used for any purpose other than personal use. Therefore, reproduction, modification, storage in a retrieval system or retransmission, in any form or by any means, electronic, mechanical or otherwise, for reasons other than personal use, is strictly prohibited without prior written permission.