* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download 1 - Cal Poly
Genomic library wikipedia , lookup
Neuronal ceroid lipofuscinosis wikipedia , lookup
Saethre–Chotzen syndrome wikipedia , lookup
Copy-number variation wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
X-inactivation wikipedia , lookup
Metabolic network modelling wikipedia , lookup
Genetic engineering wikipedia , lookup
Gene expression profiling wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Gene desert wikipedia , lookup
Genome evolution wikipedia , lookup
Genome (book) wikipedia , lookup
Gene therapy wikipedia , lookup
Gene expression programming wikipedia , lookup
Point mutation wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
History of genetic engineering wikipedia , lookup
Gene nomenclature wikipedia , lookup
Genome editing wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Helitron (biology) wikipedia , lookup
Microevolution wikipedia , lookup
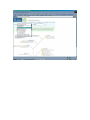
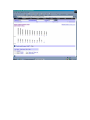
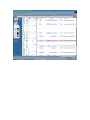
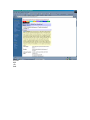
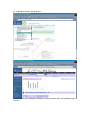
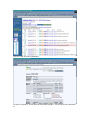
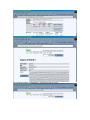
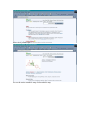
1.) Open your Web Browser such as Internet Explorer and type in the following address http://www.ncbi.nlm.nih.gov To Search for information on DNA you have sequenced 1.) From the above website choose BLAST which is in the toolbar above the search box. 2.) You will then need to choose the appropriate databases to search depending on what type of DNA you have sequenced. (ie, genomic, translated proteins, etc) In this example I have chosen to search the nucleotide-nucleotide (blastn) 3.) Once you have chosen your database you will be given search options. 4.) Copy your DNA sequence and Paste it into the Search box. 5.) Choose your appropriate database (nr will search all databases) 6.) Hit BLAST! 7.) Your search is put into a queue and estimated time is given for the completed search. 8.) Hit Format! to see your results 9.) A list of results will be given with a score and E value to show how close your sequence was to the blast results. 10.) Click on the link of your results for a detailed description. Finding the physical location of your gene 1.)Return the www.ncbi.nlm.nih.gov homepage and follow the Map Viewer option under the Hot Spots. This will lead you to http://www.ncbi.nlm.nih.gov/mapview/ 2) Either select your organism of choice (eg. S. cerevisiae) from the search pull down menu or click on the name of the organism in the phylogenetic tree. If you are looking for a specific gene you can type the name of it in the search box to the right of the pull down menu (eg. act1). 3) This will lead you to a map of the chromosomes of the organism. 4) If you searched for a specific gene, a red mark will appear next to any chromosomes which contain a matching gene. Click on this chromosome or other chromosome of interest to view physical location. Or click on gene under search results corresponding to the gene of interest. 5) Zoom out too see surrounding genes. 6) Click on gene of interest (ACT1) for more information. Finding a gene in the human genome 1) Follow instructions above to get to map viewer. www.ncbi.nlm.nih.gov/mapview/ ALDH2 hbb cftr dmd eg. arabidopsis formate dehydrogenase click on chromosome with gene of choice or on map element link corresponding to gene of choice. click on TAIR for more information to retrieve sequence: scroll down to nucleotide sequence. select data you are interested in. for enzymatic pathway information: click tools at top of screen select AraCycPathways too see the entire metabolic map click metabolic map too see pathway involving a specific enzyme click main Query Page enter protein of interest in the query by name entry field eg. formate dehydrogenase select reaction of choice select the in pathway link moving the mouse across the blue lines will give you information about the reaction. moving the mouse across the red text will give you more information about the compound in the lower left corner of the page.