* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Finding Sequences to Use in Activities
Primary transcript wikipedia , lookup
Zinc finger nuclease wikipedia , lookup
Genome (book) wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Gene expression programming wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Transposable element wikipedia , lookup
Copy-number variation wikipedia , lookup
Gene expression profiling wikipedia , lookup
Genomic library wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Human genome wikipedia , lookup
Gene therapy wikipedia , lookup
Non-coding DNA wikipedia , lookup
Genetic engineering wikipedia , lookup
Pathogenomics wikipedia , lookup
DNA barcoding wikipedia , lookup
Gene nomenclature wikipedia , lookup
Gene desert wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
History of genetic engineering wikipedia , lookup
Point mutation wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Genome evolution wikipedia , lookup
Metagenomics wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Designer baby wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Genome editing wikipedia , lookup
Microevolution wikipedia , lookup
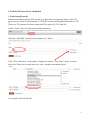
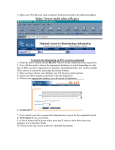
Finding Sequences to Use in Activities DNA sequences are deposited into a database known as GenBank, which is accessible through the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov) This is what the NCBI Home Page looks like: In order to pull useful sequences, you need to have some idea of what you are looking for. Let’s use the chimpanzee (Pan troglodytes) sequence from our DNA bracelet activity as an example: GTATTTGTGGTAAACCCAGTG In the bracelet activity, it was given to you as a chimpanzee sequence. So let’s BLAST it and find out what it is. Go to BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) (you can get there by clicking on the BLAST link on the right hand navigation bar on the NCBI homepage). 1 You have a nucleotide sequence, so choose nucleotide blast (right beneath the Basic BLAST)that will bring you to this page: 2 Type the chimpanzee bracelet sequence into the “Entry Query Sequence” Box (at the top). Now click on BLAST (at the bottom) and wait for your results (which will look like this): The red (thick) line at the top represents the sequence you entered. The colored lines below (blue and black) represent “matching” sequences. Scroll down to see the sequences represented by each “match”. The top listing corresponds to the top blue bar, the 2nd listing corresponds to the 2nd blue bar and so on. 3 As you can see, there are several perfect (100% matches) for this short sequence. All of these are for the gene called “granulysin”- this suggests that the sequence you entered is part of a granulysin gene. If you would like to learn more about granulysin, try using the OMIM website (part of NCBI)- go back to the NCBI homepage, select OMIM from the search menu at the top, and then type in granulysin. You can also try a general internet search. If we go back to your granulysin BLAST matches, you can see that you have “hit” granulysins from a variety of species. If you click on each match from the listing, you will get an expanded view (with the two sequences aligned). From here, you can cut and paste the species name into a search engine to see what species your segment of a granulysin gene matched [e.g. sooty mangabey- an old world monkey, orangutan, chimpanzee…]. Clearly this portion of this gene is not unique to chimpanzees. But it does appear to consistently “hit” granulysin. So this might be a good sequence if you wanted students to know that a specific gene was present, but not necessarily a specific organism (species). 4 This brings us (finally) to the question of finding appropriate sequences. It will depend on what you are trying to accomplish. Do you want students to distinguish between pathogenic (disease causing) E. coli and non-pathogenic E. coli? In this case, you may want to use the sequence for the shiga-like toxin made by many strains of pathogenic E. coli. The presence or absence of this sequence (in combination with E. coli) will tell students whether or not the E. coli is toxigenic. On the other hand, you may want students to use DNA sequences to identify specific organisms (e.g. is there E. coli and/or Staphylococcus aureus present? e.g. is that cheese made from cow’s milk, sheep’s milk, goat’s milk or camel’s milk?). In these cases, you will need to find sequences that unambiguously “hit” just the organism of interest. In the cheese example, if you were to pick a gene for a major milk protein (e.g. casein), you might not be able to tell the organisms apart, because they all have a casein gene (and the differences between organisms may not be enough to uniquely identify them- much as the segment of the granulysin gene wasn’t enough to distinguish between a variety of related primates). So what sequences can we start with to distinguish individual organisms (species)? PROKARYOTES Start with a gene called the 16S rRNA gene. This is the same gene that Carl Woese used to generate the three-domain tree of life. The gene (DNA sequence) encodes an RNA molecule that is part of the ribosome. All cellular organisms have ribosomes (to make proteins), so it is a great molecule to compare between organisms. The “S” stands for “Svedberg”, a unit that represents how fast sedimentation occurs for a molecule. The rate at which a molecule settles provides information as to its size. EUKARYOTES A. You can use a gene called the 12S rRNA gene. This is the eukaryotic equivalent of the bacterial 16S rRNA gene- the same one Carl Woese used to generate the three-domain tree of life. Because this gene evolves very slowly, there may not be many (or any) differences in the 12S rRNA gene between closely related species. In this case, you might want to try a slightly more variable gene/DNA region. B. DNA barcodes (potentially useful for closely related species) Barcode sequences are specific sequences that have been used to distinguish and identify eukaryotic organisms (particularly ones that may be so closely related that there may not be many –or any- differences in their 12S rRNA genes). They are called barcodes because, just like UPC scan codes on consumer products allow specific recognition of that product, DNA barcodes specifically identify individual species. 5 Which barcode you use will depend on the specific organism: For ANIMALS, we use a region of a mitochondrial gene called CO1 (cytochrome c oxidase subunit 1) For PLANTS, we use a region of a chloroplast gene called rbcL (rubisco large subunit) For FUNGI, we use a region of DNA called ITS (the nuclear internal transcribed spacer- a region that surrounds a particular rRNA gene) See http://www.dnabarcoding101.org/introduction.html for more information on DNA barcoding. Let’s Practice: Find each of these sequences (below). For each, you will want to save two things: a. the Accession number (this is the number permanently linked to this sequence, and will allow you to find the same sequence again) b. the FASTA sequence format (this is the format that your students will use to cut and paste into BLAST during an activity) 1. Find the 16S rRNA gene sequence for E. coli Start at the NCBI homepage. Drop down the search menu to “gene” (as you are looking for the 16S rRNA gene). Type in 16S rRNA Escherichia coli… Look through your hits for something that is just the rRNA gene: The 2nd hit looks good- click on it: 6 Now click on the FASTA link: At this point: 7 Note the NCBI Reference Sequence (the accession number for this sequence) Copy and Paste everything from the “>” to the last nucleotide into a new Word document. Now shorten its name (keep the “>” but call it something like “>E coli 16S rRNA”). SAVE it. Now take a moment to actually look at it. You can see that the page title is the “complete genome” of this strain of E. coli. But the entire genome would be enormous (you would still be waiting to finish downloading it). So this is actually the region from nucleotide 4035531 to 4037072 in the E. coli genome. This is the region that encodes the 16S rRNA gene. As a math exercise, you could ask your students to figure out how long this gene is, and what % of the E. coli genome it represents (the entire genome is approximately 5 million base pairs in length). 2. Now try to find the 12S rRNA for a cow (make sure to get the accession number and the FAST format sequence) Hint: you may want to search under “nucleotide” as well as “gene”, use 12S rRNA and the scientific name for a cow. Remember that when you are reviewing your hits, make sure that you not clicking on something else (like a mitochondrial RNA chaperone). If there are several possible hits, generally speaking, the longer sequence will probably work better. Note the Accession Number Copy and Paste the FASTA sequence and shorten its title. 3. Now try to find the 12S rRNA for a pig (get the Accession Number and the FASTA format sequence) 4. Find the DNA barcode for a cow [Search from the NCBI homepage, using the nucleotide dropdown menu; type in “cow barcode”] Make sure you note the Accession number and copy the FASTA format sequence 8 5. Find the DNA barcode for a dandelion 6. Find a fungal barcode Internal transcribed spacers (ITS) are the go-to barcodes for comparing fungi, since COI genes vary too much to be informative. 5.8S rRNA are the small segment between two ITS. These two ITS separate the three conserved rRNA genes 18S, 5.8S, and 26S. Under “Gene”, select the advanced search parameters: Click the “All Fields” dropdown and change it to “Filter”: Click “Show index list” on the right to display the options. Then select “fungi” from the index list. Then in the second query box type “internal transcribed spacer”: Your results will look like this: 9 Choose a sequence. “Saccharomyces cerevisiae” is a species of yeast using in baking and brewing, whereas “Chaetormium thermophilum” is a heat-loving fungus that grows on dung and compost. As you can see, the builder tool allows you to optimize your searches based on what you already know and what you want to find out. There are numerous options to choose from without even having to name a specific organism. These advanced search tools are found in many of the NCBI databases and help you to quickly narrow down your search without having to know the syntax for doing advanced searches. 7. Find a sheep casein gene Copy the FASTA formatted sequence (just the nucleotides, not the “> title line”) Now go to BLAST and do a nucleotide BLAST of the sheep casein gene. Did you hit the casein gene of any other species? (Hint: you can click on the “Taxonomy Reports” link near the top of the page to get a list of species that you hit) 10 11