* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download emboj2008205-sup
Genealogical DNA test wikipedia , lookup
DNA damage theory of aging wikipedia , lookup
DNA profiling wikipedia , lookup
United Kingdom National DNA Database wikipedia , lookup
Metagenomics wikipedia , lookup
Transposable element wikipedia , lookup
Copy-number variation wikipedia , lookup
DNA vaccination wikipedia , lookup
Neocentromere wikipedia , lookup
X-inactivation wikipedia , lookup
Non-coding DNA wikipedia , lookup
Nucleic acid double helix wikipedia , lookup
Molecular Inversion Probe wikipedia , lookup
Molecular cloning wikipedia , lookup
Pathogenomics wikipedia , lookup
DNA supercoil wikipedia , lookup
Designer baby wikipedia , lookup
Epigenomics wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
SNP genotyping wikipedia , lookup
History of genetic engineering wikipedia , lookup
Gel electrophoresis of nucleic acids wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Microevolution wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
Comparative genomic hybridization wikipedia , lookup
Point mutation wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Genomic library wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Cell-free fetal DNA wikipedia , lookup
Bisulfite sequencing wikipedia , lookup
Helitron (biology) wikipedia , lookup
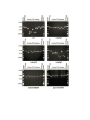
Materials and methods Strains and Plasmids All strains in this study were isogenic to KT119 strain (MATa, his7-2, leu2-3,112, trp1-, ura3-, lys2-, ade2-, bar1-, sfa1-, cup1-1-, yhr054c- cup1-2-, lys2::kanMXURA3, ADE2, CUP1, SFA1derivative of TP strains described in Narayanan et al., 2006. GAA/TTC repeats of length 20, 60, 120 and 230 were integrated into LYS2 in two orientations using the dellito perfetto technique (Storici et al., 2001). GAA repeats located on the plasmids (Krasilnikova and Mirkin, 2004) were used as the source for PCR amplification. Strains with 340 repeats in both orientations were a result of natural expansions from 230 repeats. The length of the repeat tracts and the absence of interruptions were determined by sequencing from both ends. MMR genes were disrupted with the kanMX cassette (Wach et al., 1994). msh2-G693A and pms1-E707K alleles were introduced using dellito perfetto technique. Nucleotide sequences of the primers used for integrations and disruptions are available upon request. Genetic Techniques The rates and 95% confidence intervals of the arm loss and recombination between lys2 alleles were estimated in fluctuation tests using at least 14 independent cultures (Lobachev et al., 1998). Single colonies taken into the test were prescreened for the presence of full-sized tracts using colony PCR. The canavanine-containing media was made with a low concentration of adenine (5mg/L) to allow color detection. Structural Analysis of the Genome Rearrangements Chromosome aberrations were characterized using CHEF (contour-clamped homogeneous electric field) gels and Southern Blot Hybridization with MET6-specific probe as previously described (Narayanan et al., 2006). Genomic DNA preparation and subsequent CGH microarray analysis of the CanRAde- isolates were performed according to procedures described (Lemoine et al., 2005). Arrays were analyzed using GenePix pro 4.1 (Axon Instruments) and Gene Spring ® 5.1 (Silicon Genetics). A complete analysis of the microarrays can be found on line at GEO database (accession number GSE11425). The translocation breakpoints were PCR amplified and sequenced with primers designed based on microarray data. For DSB detection, chromosomal DNA was embedded into agarose plugs using the CHEF Genomic DNA plug Kit from Bio-Rad (~6 x 108 cells/1ml of plug). Gels were run in 0.5X TBE at 14°C using the Bio-Rad CHEF Mapper XA for 19.44 hours with switch times of 2.08s-42.91s. Southern blot hybridization was performed using 350 bp probe homologous to DSF1 gene as previously described (Lobachev et al., 2002). 2D Analysis of Replication Fork Intermediates Cells were arrested in G1 with -factor and released synchronously into S-phase. At 40 min after release, chromosomal DNA was extracted according to (Friedman and Brewer, 1995). Neutral/neutral 2-D analysis was carried out in according to (Brewer and Fangman, 1987). The AflII-digested DNA was separated in the first dimension on a 0.4% gel without ethidium bromide in 1X TBE buffer at 1V/cm for 38 hours in first dimension. The second dimension gel was run at 6 V/cm in 1X TBE buffer containing 0.3 g/ml ethidium bromide for 13 hours. Southern blot hybridization using 4 kb AflII-digested LYS2 fragment was performed to highlight replication intermediates. Supplemental References Brewer, BJ, Fangman, WL (1987) The localization of replication origins on ARS plasmids in S. cerevisiae. Cell 51: 463-471. Friedman, KL, Brewer, BJ (1995) Analysis of replication intermediates by two-dimensional agarose gel electrophoresis. Methods Enzymol. 262: 613-627. Krasilnikova, MM, Mirkin, SM (2004) Replication stalling at Friedreich's ataxia GAAn repeats in vivo. Mo.l Cell. Biol. 24: 2286-2295. Lemoine, FJ, Degtyareva, NP, Lobachev, K, Petes, TD (2005) Chromosomal translocations in yeast induced by low levels of DNA polymerase a model for chromosome fragile sites. Cell 120: 587-598. Lobachev, KS, Gordenin, DA, Resnick, MA (2002) The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell 108: 183-193. Lobachev, KS., Shor, BM, Tran, HT, Taylor, W, Keen, JD, Resnick, MA, Gordenin, DA (1998) Factors affecting inverted repeat stimulation of recombination and deletion in Saccharomyces cerevisiae. Genetics 148: 1507-1524. Narayanan, V, Mieczkowski, PA, Kim, HM, Petes, TD, Lobachev, KS (2006) The pattern of gene amplification is determined by the chromosomal location of hairpin-capped breaks. Cell 125: 1283-1296. Storici, F, Lewis, LK, Resnick, MA (2001) In vivo site-directed mutagenesis using oligonucleotides. Nat. Biotechnol. 19: 773-776. Wach, A, Brachat, A, Pohlmann, R, Philippsen, P (1994) New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10: 1793-1808. Supplementary Table I. CanRAde- isolates of strains with (TTC)230 and (GAA)230 repeats Isolate T-1, T-3, T-4, T-5, T-8, T-11, T-12 T-7, T-9 T-6 T-2, T-10 Rearrangement deletion+ non reciprocal translocation (Left arm, Chr.I), 7/12 (58%)a deletion+non reciprocal translocation (Left arm, Chr.XIII), 2/12 (17%) deletion+non reciprocal translocation (Left arm, Chr.II), 1/12(8%) deletion+telomere addition, 2/12 (17%) A-1,A-2, A-3, A-5, A-6, A-7, A-9, A-10, A-11, A12 deletion+non reciprocal translocation (Left arm, Chr.XI), 10/12 (83%) A-4, A-8 deletion+non reciprocal translocation (Left arm, Chr.XIII), 2/12 (17%) Breakpoint ORF GAA/TTC-rich sequence used for invasion and BIRb FUN12 CTTCGTTAGCTTC←TTCTTCGTTCTT←T TCTTCCAATAACTTCTCTTCTTCCTACC ATTCGGATATGATTTCAGGTTCTTCCTT TTCCTCTTCCTTTTTTTCCTTTTCCTCTT CGTCTTCTTCTTTCTT ORC1 CTCCTTCTTCTTCTGCTTCT←TCTTCTG CTTCTGCTTCTTCTCTTTCTT SCT1 TCCCTTCTCCTTCTCCTTCTTCTTCTCCT TCTTC←TTCTTCTCCTTCTTCTTCTTCTT CCCTTTCTTCCTC - - MNN4 GAGGAGGACGAAGAAAAAAAGG←AG AAGAAGCAAGAAGAAGAAAAGAAGGA GAAGGACGAAGAAGAAAAGAAGTAGA AGTAAGAAGAACAAAAAGAGAAGGAC GAAGAAAAAAAGGAGAAGAAGCAAGA AGAAGAAAAGAAGTAGAAGTAAGAAG AATAAAAGAAGTAGAAGTAAGAAGTA GAAAAGAGGAAGAAGGACGAAGAAGA AGA←GAAGAAGAAGGAAGAAGAA←A AAGAGGAGAAGGAGGACGAAGAAGAA AAGAAGAAGAAGGAAGAAAAAGAAGA GAAGAAGAAGGAAGAAAAAGAAGAGA AGGAGAAGGAAGAAAAAGAAAAGAAG AAGAAGGAAGAAGAAGAAAAGGAGAA GAAGGAAGAAGAAGAAGAGGAGAAGG AGGAAGAAGAAGAAGAGGGAGGCGAA AGAAAGAAG FPR3 a The percentage of isolates that belong to the GCR GAAGAAATGAAAAAATCGAAAGAAAA GAAGAAGAA←AGAGAA←GCACGAAA GAGAAAAAAAGGAAAAACGAAAAGAA TCCAAGACCGAATTGAAGAAGAAAAA CAAGAAGAAGAAGAAGAAG AAGAACAAGAAGAAGAGGTAGTAATA class GAAGAAGTAGAAGAAGAACCTGAAGA is indicated in parentheses. A b Blue and red arrows indicates the point of junction with the invaded TTC or GAA strands from chromosome V, respectively. Multiple arrows indicate invasion point identified for independent isolates Supplementary Table II. Length- and orientation-dependent size variations in expanded GAA and TTC repeat tracts. Insertion in LYS2 (GAA)60 Number of colonies examined 120 Contraction* (%) Expansion* (%) Unchanged (%) <1 <1 100 (GAA)230 (GAA)340 (TTC)60 (TTC)230 120 120 60 120 3.3 88.3 <1.7 2.5 1.7 5.8 <1.7 1.7 95 5.8 100 95.8 (TTC)340 120 4.2 3.3 92.5 * The single colonies verified for the presence of full-size repeat tracts were re-streaked on YPD media and size changes in the progeny colonies were determined by PCR analysis on genomic DNA (20-50 ng was used as template). Colonies derived from three independent isolates for each tract length were examined. Expansions and contractions of the repeats were scored if the PCR analysis revealed the presence of a novel band different from the original repeat length. In most of the cases, the PCR products contained only one discrete band. Supplementary Table III. Chromosomal arm loss events in wild type and MMR mutants containing (GAA)230 tracts. Genotype Wild type Δmsh2 Δmsh3 Δmsh6 Δmsh3Δmsh6 Δpms1 Δmlh1 msh2-G693A pms1-E707K Arm loss rate (x 107)a 66 (51-86)b 4 (2-9) 12 (11-17) 42 (31-51) 6 (2-7) 4 (2-5) 5 (1-6) 6 (2-8) 5 (2-8) Fold reduction over wild type 1 16 6 1.6 11 16 13 11 13 a The loss of CAN1 and ADE2–containing region was measured in strains that do not have the lys2-8 allele. b Numbers in parentheses correspond to the 95% confidence interval Supplementary figure legends Figure 1. Effect of mutations in MMR on (GAA)340 tract stability. To determine the frequency of expansions and contractions of repeat tracts during mitotic divisions, we re-streaked yeast colonies that have been verified for the presence of (GAA)340 full size repeats on complete media. Ten colonies were then selected for PCR amplification to look for changes in the length of the repetitive tracts. The lanes labeled with asterisks denote unchanged tracts length. The band on the lane labeled (GAA)340 is 1389 bp and corresponds to the full-size repeats plus flanking sequences. The primer-pair that reliably worked in colony PCR reaction was determined experimentally. Analysis of tract length variations in mlh1, pms1, msh2-G693A and pms1-E707K strains yielded results similar to those observed in msh2 and msh3 strains.