* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download HIV Drug Resistance: What We Know and How We Know It
Neuropharmacology wikipedia , lookup
Drug interaction wikipedia , lookup
Discovery and development of HIV-protease inhibitors wikipedia , lookup
Pharmaceutical industry wikipedia , lookup
Drug design wikipedia , lookup
Drug discovery wikipedia , lookup
Prescription costs wikipedia , lookup
Pharmacogenomics wikipedia , lookup
Pharmacokinetics wikipedia , lookup
Discovery and development of integrase inhibitors wikipedia , lookup
Discovery and development of non-nucleoside reverse-transcriptase inhibitors wikipedia , lookup
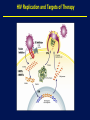
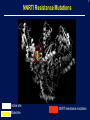
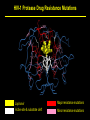
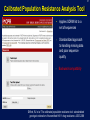
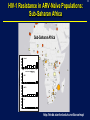
1 Rationale and Uses For a Public HIV Drug Resistance Database Bob Shafer, MD Professor of Medicine and by Courtesy Pathology (Infectious Diseases) 2 Outline • HIV drug therapy essentials • HIVDB • Examples of public health applications • Surveillance of transmitted drug resistance • Genetic mechanisms of acquired drug resistance HIV-1 Genome HIV Replication and Targets of Therapy 5 Antiretroviral Inhibitors (ARVs) ddI ddC AZT 1990 Nucleoside RT Inhibitor 3TC d4T NVP IDV RTV SQV EFV DLV APV NFV ABC 1995 LPV TDF 2000 Protease Inhibitor Nonnucleoside RT inhibitor T20 MVC ATV FTC TPV DRV RAL ETR 2005 Fusion Inhibitor Integrase Inhibitor CCR5 Inhibitor 6 HIV Genetic Variation • Generation of variation • High mutation rate • Recombination • Proviral DNA “archive” • Selective evolutionary pressures • Immunological • Antiretroviral drugs (ARVs) Tebit DM, Arts EJ. Tracking a century of global expansion and evolution of HIV. Lancet Infect Dis 2011 8 HIV-1 RT: Active Site, Template, Primer, and dNTP Active site Incoming nucleotide 9 NNRTI Resistance Mutations Active site Etravirine NNRTI resistance mutations HIV-1 Protease Drug Resistance Mutations Lopinavir Major resistance mutations Active site & substrate cleft Minor resistance mutations Models Relating HIV Drug Resistance to Treatment Response 12 10 Million Patients on Antiretroviral Therapy 2013 Global AIDS Response Progress Reporting (WHO/UNICEF/UNAIDS) 13 Outline • HIV drug therapy essentials • HIVDB • Examples of public health applications • Surveillance for transmitted drug resistance • Genetic mechanisms of acquired drug resistance 14 Database Rationale • Drug resistance knowledge important for Interpreting genotypic resistance tests Designing surveillance studies and public health decisions Assisting drug development. 15 How we know what we know about HIV drug resistance mutations • Genotype-treatment correlations – 1998 • Genotype-phenotype correlations – 2002 • Genotype-outcome correlations – 2005 16 Database Rationale • Large amounts of drug resistance data are important for generating drug-resistance knowledge. • Uniform representation of 3 main data correlations facilitates meta-analyses. Genotype-Rx Genotype-Phenotype Genotype-Outcome Clinical management Epidemiologic studies Drug development http://hivdb.stanford.edu Genotypic HIV Resistance Testing CCTCAGATCACTCTTTGGCAACGACCCATAGTCACAATAAAGATAGCGGGACAACTAAAGGAAGCTCTATTAGATACAGGAGCAGATGATACA GTATTAGAAGAAATGAATTTGCCAGGAAAATGGAAACCAAAAATAATAGTGGGAATTGGAGGGTTTACCAAAGTAAGACAGTATGATCATGTAC AAATAGAAATCTGTGGACATAAAGTTATAGGTGCAGTATTAATAGGACCTACACCTGCCAATATAATTGGAAGAAATCTGTTGACTCAGCTTGGC TGTACTTTAAATTTT PQITLWQRPIVTIKIAGQLKEALLDTGADDTVLEEMNLPGKWKPKIIVGIGGFTKVRQYDHVQIEICGHKVIGAVLIGPTPANIIGR NLLTQLGCTLNF Differences from Consensus B: L10I, G17R, K20I, E35D, N37S, M46I, I62V, L63P, A71I, G73S, I84V, L90M, I93L HIV-1 Genotypic Resistance Testing: Online Interpretation Meaningful Results (1) Quality control (2) Sequence Interpretation (3) Literature references (4) Clinical education / advice Shafer RW et al. HIV-1 RT and Protease Search Engine for Queries. Nat Med 2000 HIVdb: Genotypic Resistance Interpretation http://hivdb.stanford.edu HIVdb: Genotypic Resistance Interpretation HIVdb: Genotypic Resistance Interpretation HIVdb: Genotypic Resistance Interpretation 24 Surveillance for Transmitted Drug Resistance 25 Outline • HIV drug therapy essentials • HIVDB • Examples of public health applications • Surveillance for transmitted drug resistance • Genetic mechanisms of acquired drug resistance 26 Rationale for Surveillance for Drug Resistance in ARV-Naive Populations • Assess extent of transmitted drug resistance (TDR). • Monitor the expected efficacy of first-line therapies. 27 Challenges to ARV-Resistance Surveillance • There is no perfect definition of genotypic resistance. • There are many different drug-resistance mutations (DRMs). • Drug resistance mutations occasionally occur in the absence of selective drug pressure. Therefore, not all drug-resistance mutations are evidence for transmitted drug resistance (TDR). 28 Challenges to ARV-Resistance Surveillance • More than 300 studies of genotypic resistance in ARVnaïve patients have been published. • Findings differ by region, time, study population, and potentially study methods. 29 Surveillance Drug Resistance Mutations (SDRMs) • Drug-resistance mutations with a high sensitivity and specificity for detecting selective ARV pressure. • Nonpolymorphic. • Applicable to all HIV-1 subtypes. Shafer RW, et al. HIV drug resistance mutations for drug resistance surveillance. AIDS 2007 30 HIV-1 Resistance in ARV-Naïve Populations: Analysis of Published RT and PR Sequences • Well-characterized representative population of ARVnaïve persons. • Country and year of virus isolation known. • HIV-1 RT ± PR sequence is publicly available. 31 Calibrated Population Resistance Analysis Tool • Applies SDRM list to a set of sequences • • Standardized approach to handling missing data and poor sequence quality. Backward-compatibility Gifford, RJ et al. The calibrated population resistance tool: standardized genotypic estimation of transmitted HIV-1 drug resistance. AIDS 2008 HIV-1 Resistance in ARV-Naïve Populations: Prevalence by Region Region No. Studies No. Persons % Resistance Median % Resistance IQR North America 24 11,038 11.4 8.8 – 14.0 Europe 44 11,419 9.3 6.0 – 15.1 Latin America 39 5,802 7.6 4.0 – 10.1 High-income Asia 11 3,190 5.5 3.5 – 9.0 Former Soviet Union 11 1,124 3.4 0.0 – 6.4 South/Southeast Asia 49 4,181 3.3 2.0 – 5.3 Sub-Saharan Africa 86 9,904 2.8 1.1 – 5.7 264 46,660 HIV-1 Resistance in ARV-Naïve Populations: Sub-Saharan Africa Sub-Saharan Africa Overall 8 4 0 887 1529 <=1 1538 <=3 1343 4 1803 <=6 1200 7 1449 <=14 %Resistance Years Since AR NRTI 8 4 0 NNRTI 8 4 0 PI 8 4 0 http://hivdb.stanford.edu/surveillance/map/ 33 HIV-1 Resistance in ARV-Naïve Populations: South / Southeast Asia South/Southeast Asia Overall 8 4 0 495 512 858 556 877 811 <=2 3 4 <=6 <=9 %Resistance Years Since AR NRTI 8 4 0 NNRTI 8 4 0 PI 8 4 0 http://hivdb.stanford.edu/surveillance/map/ 34 HIV-1 Resistance in ARV-Naïve Populations: Most Common SDRMs by Region and ARV Class 35 HIV-1 Resistance in ARV-Naïve Populations: Conclusions • Significant differences in prevalence of resistance in ARVnaïve patients by region and year. • Transmitted NNRTI resistance is increasing in SubSaharan Africa and South/Southeast Asia. • Analysis of data from many studies is required to obtain meaningful estimates of transmitted drug resistance. 36 37 Outline • HIV drug therapy essentials • HIVDB • Examples of public health applications • Surveillance for transmitted drug resistance • Genetic mechanisms of acquired drug resistance 38 Rationale • In resource-limited regions, ~25% of patients receiving first-line ART develop virological failure within 1 year. • Drug-resistance mutations are detected in 50% to 90% of patients with virological failure. • Regimens used in resource-limited countries differ from those used in well-resourced countries. • Patients in resource-limited countries are monitored infrequently and second-line therapy is chosen without genotypic resistance testing. 39 Genetic Mechanisms of Resistance in Patients with Virological Failure • Choosing second-line therapy. • Developing point-of-care (POC) diagnostic tests. 40 WHO-Recommended First-Line ARV Regimens WHO-Recommended Regimens, 2016 to 2013 NRTI NRTI NNRTI / PI d4T (being phased out) 3TC (or FTC) EFV AZT NVP TDF LPV (PI, 2nd line) ABC (children) 41 Number of Patients by Regimen and Subtype Data summary from mid 2012 A B C AE AG D G Misc Total d4T/3TC/NVP 50 55 121 430 123 27 122 40 1121 AZT/3TC/NVP 45 99 394 45 50 42 46 21 469 d4T/3TC/EFV 13 92 188 16 9 2 3 11 540 AZT/3TC/EFV 25 244 274 45 20 17 26 11 576 133 490 977 536 202 88 197 83 2706 42 Sources of Patient Data and Sequences Data summary from mid 2012 Number Studies Number Patients % 10 largest 1,409 51% 20 largest 1,981 72% 50 largest 2,640 98% 43 Question From WHO: Which NRTI should be substituted in patients stopping d4T? • Patients with virological failure on d4T can develop resistance by two mutually exclusive mutational pathways: • Thymidine analog mutations: cross-resistance to AZT • Non-thymidine analog mutations particularly K65R: cross-resistance to TDF and increased susceptibility to AZT • In vitro studies have shown that viruses belonging to subtype C are at increased risk for developing K65R. 44 Impact of NNRTI, Subtype, and Years on NRTIResistance Mutations in 1,840 Patients Receiving d4T 45 Impact of Subtype on AZT and TDF Cross-Resistance in 1,840 Patients Receiving d4T 46 Rationale for Point-Of-Care (POC) Resistance Testing in Low/Middle-Income Countries? • POC test for detecting virological failure have been developed. • A POC resistance test for a limited number of the most important mutations could be used: • To confirm virological failure • To suggest among second-line therapy options • Be used prior to therapy in regions with elevated TDR or in patients with uncertain treatment history. 47 Sensitivity for Detecting Resistance after 1st-Line Failure: 4 NNRTI and 6 NRTI-Resistance Mutations 48 Sensitivity for Detecting Resistance in Untreated Patients: 4 NNRTI and 6 NRTI-Resistance Mutations 49 Conclusions • Drug resistance knowledge is important for interpreting genotypic resistance tests, designing surveillance studies, and drug development. • Large amounts of drug resistance data are important for generating drug-resistance knowledge. • Drug-resistance data consists mostly of correlations between genotype-treatment, genotype-phenotype, and genotypevirological outcome. 50 Acknowledgements Database / Data analysis Soo-Yon Rhee, M.S. Tommy Liu, B.S. Michele Tang, M.D. Vici Varghese, Ph.D. Funding NIAID – Division of AIDS Bill and Melinda Gates Foundation 51 HIV-1 Evolution and Drug Resistance: An Example A B HIV-1 levels prior to TMB-202 HIV-1 levels during and following TMB-202 June 2009 2009 April 2010 Plasma HIV-1 RNA log copies / ml 1997 6.0 5.0 ENF 4.0 DRV + RAL 3.0 EFV 2.0 Below the level of quantification Below the level of quantification 1.0 97 98 99 00 01 02 03 04 05 06 07 08 09 -4 0 4 8 12 16 20 24 28 32 36 40 44 Ibalizumab Infusions Accompanying antiretrovirals: etravirine + enfuvirtide Fessel WJ, et al. The efficacy of an anti-CD4 monoclonal antibody for HIV-1 treatment. Antivir Res 2011