* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download PDF
Survey
Document related concepts
Transcript
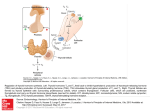
0888-8809/01/$03.00/0 Printed in U.S.A. Molecular Endocrinology 15(9):1529–1538 Copyright © 2001 by The Endocrine Society Aberrant Alternative Splicing of Thyroid Hormone Receptor in a TSH-Secreting Pituitary Tumor Is A Mechanism for Hormone Resistance SHINICHIRO ANDO, NICHOLAS J. SARLIS, JAY KRISHNAN, XU FENG, SAMUEL REFETOFF, MICHAEL Q. ZHANG, EDWARD H. OLDFIELD, AND PAUL M. YEN Molecular Regulation and Neuroendocrinology Section (S.A., N.J.S., J.K., X.F., P.M.Y.), Clinical Endocrinology Branch, National Institute of Diabetes and Digestive and Kidney Diseases, and Surgical Neurology Branch (E.H.O.), National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, Maryland 20892; Departments of Pediatrics and Medicine (S.R.), University of Chicago, Chicago, Illinois 60637; and Cold Spring Harbor Laboratory (M.Q.Z.), Cold Spring Harbor, New York 11724 Patients with TSH-secreting pituitary tumors (TSHomas) have high serum TSH levels despite elevated thyroid hormone levels. The mechanism for this defect in the negative regulation of TSH secretion is not known. We performed RT-PCR to detect mutations in TR from a surgically resected TSHoma. Analyses of the RT-PCR products revealed a 135-bp deletion within the sixth exon that encodes the ligand-binding domain of TR2. This deletion was caused by alternative splicing of TR2 mRNA, as near-consensus splice sequences were found at the junction site and no deletion or mutations were detected in the tumoral genomic DNA. This TR variant (TR2spl) lacked thyroid hormone binding and had impaired T3-dependent negative regulation of both TSH and glycoprotein hormone ␣-subunit genes in cotransfection studies. Furthermore, TR2spl showed dominant negative activity against the wild-type TR2. These findings strongly suggest that aberrant alternative splicing of TR2 mRNA generated an abnormal TR protein that accounted for the defective negative regulation of TSH in the TSHoma. This is the first example of aberrant alternative splicing of a nuclear hormone receptor causing hormonal dysregulation. This novel posttranscriptional mechanism for generating abnormal receptors may occur in other hormone-resistant states or tumors in which no receptor mutation is detected in genomic DNA. (Molecular Endocrinology 15: 1529–1538, 2001) T TRs are members of a family of nuclear hormone receptors that include the steroid hormone, vitamin D, and retinoic acid receptors (3, 4). These receptors have a variable amino terminus, a central DNA-binding domain, and a carboxy-terminal ligand-binding domain. TRs bind to their thyroid hormone response elements, usually located in the promoter regions of target genes, and regulate transcription by interacting with coactivators and corepressors (5). In positively regulated target genes, coactivators mediate hormone-induced activation, while corepressors, nuclear receptor corepressor (NCoR) and silencing mediator of retinoic acid and thyroid hormone receptor (SMRT), mediate transcriptional silencing. In negatively regulated target genes, which are not as well characterized, corepressors activate the glycoprotein hormone ␣-subunit and TSH genes, whereas coactivators such as steroid receptor coactivator-1 (SRC-1), glucocorticoid receptor interacting protein 1, thyroid receptor activator molecule 1, and activator of thyroid and retinoic acid receptors enhance the T3-dependent negative regulation of glycoprotein hormone ␣-subunit gene (6, 7). There are two distinct TR genes, TR␣ and TR. The TR␣ gene generates two proteins, TR␣1 and a car- HYROID HORMONE SYNTHESIS is tightly regulated by a negative feedback system involving the hypothalamus, pituitary, and thyroid gland. In most forms of hyperthyroidism, elevation of T3 and T4 represses secretion of TSH by the pituitary gland. However, in patients with TSH-secreting pituitary tumors (TSHomas), this negative feedback is disrupted, as TSHomas overproduce TSH in the face of elevated serum T3 and T4 levels (1, 2). Furthermore, TSH secretion cannot be stimulated by TRH. These patients typically have physical signs and symptoms of thyroid hormone excess. Additionally, TSHomas present almost invariably as macroadenomas and can cause visual and vascular complications. The mechanism for this loss of negative regulation of TSH secretion by thyroid hormone in TSHomas is not known but may involve a defect in signaling via thyroid hormone receptors (TRs). Abbreviations: GAPDH, Glyceraldehyde-3-phosphate dehydrogenase; NCoR, nuclear receptor corepressor; RTH, resistance to thyroid hormone; SMRT, silencing mediator of retinoic acid and thyroid hormone receptor; SRC-1, steroid receptor coactivator-1; ss, splice site; TIF2, transcription intermediary factor 2. 1529 1530 Mol Endocrinol, September 2001, 15(9):1529–1538 boxy-terminal splice variant, c-erbA␣2, that does not bind thyroid hormone. The TR gene generates two isoforms, TR1 and TR2, via promoter choice (8). TR1 and TR2 proteins have identical DNA- and ligand-binding domains, but differ at their amino termini. Although TR␣1, TR1, and c-erbA␣2 have widespread expression, TR2 has tissue-selective expression in the anterior pituitary gland, hypothalamus, and developing brain (9, 10). Recently, TR2-selective knockout mice have been generated, which manifest elevated serum TSH and thyroid hormone levels (11). This finding suggests that TR2 may be the critical TR isoform that negatively regulates TSH secretion. Patients with the inherited syndrome of resistance to thyroid hormone (RTH) also have elevated serum T3 and T4 concentrations and normal or elevated TSH level (12–14). These patients generally have point mutations in one of the alleles of the TR gene that results in a mutant TR that cannot bind T3 or regulate transcription but can nonetheless bind to thyroid hormone response elements of target genes. The dominant negative activity of the mutant TR on wild-type TR reduces thyroid hormone suppression of the two genes that generate TSH subunits: glycoprotein hormone ␣-subunit and TSH (15, 16). Given the precedence for TR mutations in RTH patients, we performed mutational analysis of TR isolated from a surgically resected TSHoma. Surprisingly, we detected a splice variant of TR mRNA that translates into an abnormal TR2 protein that is unable to bind T3. This abnormal TR2 was transcriptionally inactive and disrupted the negative regulation of TSH by T3 in cotransfection studies. These findings suggest that a novel, posttranscriptional mechanism caused central thyroid hormone resistance in this patient with a TSHoma. Similar posttranscriptional mechanisms may explain other hormone-resistant states or tumors in which no hormone receptor mutations can be detected from genomic DNA. Ando et al. • Aberrant Splicing of TR mRNA in a TSHoma be completely removed due to dural invasion, and the patient was started on octreotide therapy. The patient’s symptoms abated and her thyroid function tests normalized. A TRH stimulation test showed normal TSH secretory response (data not shown). A T3 suppression test performed 4 yr after surgery (Fig. 1) failed to suppress the patient’s serum TSH to less than 10% of the baseline, suggesting that at least part of the patient’s TSH secretion was due to residual tumor (1). Annual pituitary magnetic resonance imagings have shown a persistent 7-mm residual adenoma with no further growth. The patient’s clinical status has remained unchanged. Finding of TR2 mRNA Splice Variant from a TSHoma RNA from the patient’s TSHoma and pooled normal pituitaries were used to amplify full-length TR1, TR2, and an internal control, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA by RT-PCR. Similar amounts of GAPDH mRNA were detected in the samples, suggesting that similar amounts of RNA were analyzed from the TSHoma and the pooled normal pituitaries (Fig. 2A, lanes 5 and 6). Additionally, similar amounts of TR1 mRNA were measured in the TSHoma and pooled normal pituitaries (Fig. 2A, lanes 1 and 2). Surprisingly, a short variant TR2 mRNA (TR2spl) was the major amplified product from the TSHoma, as only a small amount of amplified wildtype TR2 product was observed (Fig. 2A, lane 4). In RESULTS Clinical Studies A 79-yr-old Caucasian female presented at NIH with goiter and palpitations due to hyperthyroidism. The patient had a serum TSH concentration of 10.5 U/ml (normal, 0.43–4.60), free T4 concentration of 7.4 ng/dl (normal, 0.9–1.6), and glycoprotein hormone ␣-subunit concentration of 34.1 g/liter (normal, ⬍5). Serum GH and PRL levels were normal. A magnetic resonance image of pituitary revealed an 18-mm pituitary adenoma with extension into the left cavernous sinus. Transsphenoidal surgery was performed, and immunohistochemical analysis of the tumor showed strongly positive staining for TSH and glycoprotein hormone ␣-subunit, as well as staining for both GH and PRL. Postoperatively, the patient’s serum TSH decreased to 3.1 U/ml. However, the tumor could not Fig. 1. T3 Suppression Test of the Patient T3 (300 g) was administered orally to the patient, and serum samples were obtained just before, and 48 h after, T3 administration for measurement of serum TSH. Normal basal serum TSH ranges from 0.43 to 4.60 U/liter. Normal controls suppress to less than 10% of the baseline 48 h after T3 administration. Shaded areas represent normal ranges for baseline serum TSH and expected ranges for T3-suppressed serum TSH 48 h after T3 administration, respectively. Ando et al. • Aberrant Splicing of TR mRNA in a TSHoma Mol Endocrinol, September 2001, 15(9):1529–1538 1531 Fig. 2. TR2spl Detection in a TSHoma A, RT-PCR was performed using mRNA from a TSHoma (lanes 2, 4, and 6) or pooled normal pituitary RNA from 87 individuals (lanes 1, 3, and 5). Primers were specific for the amplification of TR1 (lanes 1 and 2), TR2 (lanes 3 and 4), and GAPDH (lanes 5 and 6). Lanes 7 and 8 are products of exon 6 amplification of genomic DNA of TSHoma or circulating peripheral leukocytes of the same patient. B, Ratio of TR isoforms mRNA expression normalized with respect to GAPDH mRNA expression. contrast, only the wild-type TR2 product was amplified from the pooled normal pituitary RNA (Fig. 2A, lane 3). TR2spl mRNA was detected only in the TSHoma RNA when RT-PCR was performed using TR2 exon 6 primers (data not shown). Taken together, these findings suggested that TR2spl mRNA was specific for the TSHoma and contained a deletion within exon 6. Furthermore, since the primers used for amplification detected full-length TR2 cDNA, they enabled measurement of the relative expression of both wild-type TR2 and TR2spl mRNA within the same PCR reaction. These semiquantitative RT-PCR results showed that there is much higher expression of TR2spl mRNA than wild-type TR2 mRNA in the TSHoma, although the total amount of TR2 mRNA is not significantly different than TR2mRNA from the pooled normal pituitaries (Fig. 2B). Similar findings were observed in three repeat experiments. The intensities of the bands in Fig. 2A, lanes 3 and 4, corrected for GADPH mRNA expression, are shown in Fig. 2B. The RT-PCR products then were sequenced, and the TR2spl mRNA was found to have a 135 base in-frame deletion in exon 6 (Fig. 3). This deleted region was identical in TR1 and TR2 and encoded a part of the ligand-binding domain. Additionally, TR2spl protein had a newly created isoleucine in place of the deleted 46 amino acids in the ligand-binding domain (Fig. 3). PCR was performed using genomic DNA from 1532 Mol Endocrinol, September 2001, 15(9):1529–1538 Ando et al. • Aberrant Splicing of TR mRNA in a TSHoma by aberrant splicing via latent splice sites that existed within exon 6 of wild-type TR2 mRNA. Functional Properties of TR2 Splice Variant Fig. 3. Structure of TR2 and Location of the Deletion of TR2spl Receptor domains and the three exons encoding the ligand-binding domain of TR2 are indicated. TR2spl has a deletion from amino acids 340 to 385 substituted by an isoleucine in the ligand-binding domain. Note that exons 5, 6, and 7 of TR2 are the same as exons 8, 9, and 10 of TR1; amino acids 340–385 of TR2 are the same as amino acids 325–340 of TR1. the patient’s TSHoma and peripheral leukocytes and did not show any deletion in exon 6 (Fig. 2A, lanes 7 and 8). Analysis of Splice Sites Examination of both 5⬘- and 3⬘-sequences flanking the junction site of the deletion of TR2spl mRNA showed near-consensus splicing sequences that were located within exon 6 of wild-type TR2 (Fig. 4 A). These sequences conformed to several rules that are important for efficient splicing (17, 18). Most splice junctions are bordered by GT/AG at the 5⬘- and 3⬘-ends of the spliced sequences, as was the case for TR2spl mRNA. Additionally, there are consensus sequences for the 5⬘-splice site (ss), 3⬘-ss, and branch sequence, which are located 20 to 50 bases upstream of the 3⬘-ss. TR2 mRNA had near-consensus 5⬘- and 3⬘splice sequences, as well as a consensus branch sequence within exon 6 (Fig. 4B). Using an internal coding exon predicting program, Michael Zhang’s Exon Finder (19), the 5⬘-ss and 3⬘-ss used by TR2spl had scores indicating their high potential for alternative splicing (AAAGTGAGA, 5⬘-ss score ⫽ 46.77; TGGATGACACTGAAGTA, 3⬘-ss score ⫽75.47), although their scores were slightly lower than those of the wild-type TR ss (CAGGTGAGT, 5⬘-ss score ⫽ 49.93; ACTGGTTCTTTTCAGCT, 3⬘-ss score ⫽83.75). Aberrant alternative splicing theoretically could be due to somatic mutations in the genomic DNA of the TSHoma, which, in turn, might generate a new cryptic splice site or create an unfavorable splice sequence in the intron/exon junctions of exon 6. However, sequencing of the PCR products from genomic DNA of the TSHoma showed no mutation in exon 6 and or in the flanking intronic sequences (data not shown). These findings demonstrate that TR2spl was created Most mutations identified in patients with RTH are located in three clusters of the ligand-binding domain of TR (cluster 1: TR1 234–282, cluster 2: TR1 310–383, cluster 3: TR1 429–460) (20). The deletion of TR2spl is located within cluster 2 of the ligand-binding domain of TR2. Since most RTH mutants reduced binding to T3, the T3-binding activity of TR2spl was measured and compared with the T3-binding activities of wild-type TR2 and TR2G360R (TR2Mf), a cluster 2 mutant from a patient with RTH previously shown to have minimal T3 binding (20a). As expected from the location of the deletion in the ligand-binding domain, both TR2spl and TR2Mf showed minimal T3 binding (Fig. 5). We then examined the functional activity of the wild-type TR2 and TR2spl on the regulation of the genes encoding the two subunits of TSH: glycoprotein hormone ␣-subunit and TSH. It has been previously shown that wild-type TR can cause T3independent activation and T3-dependent negative regulation of transcription for these two genes (6, 15). As shown in Fig. 6, TR2spl lost both T3independent activation and T3-dependent negative regulation on glycoprotein hormone ␣-subunit and TSH genes. In the presence of T3, TR2spl also showed higher transcriptional activity of the glycoprotein hormone ␣-subunit and TSH genes than wild-type TR2, consistent with the clinical observation of nonsuppressible TSH in the face of elevated T3 observed in the patient (Fig. 1). Cotransfection of the wild-type TR2 with TR2spl, as well as TR2Mf, resulted in diminished T3-dependent negative regulation of both glycoprotein hormone ␣-subunit and TSH genes (Fig. 7). These results show that both TR2spl and TR2Mf have strong dominant negative activity against the wild-type TR2 on the transcription of glycoprotein hormone ␣-subunit and TSH genes. To study the interaction between TR2spl and transcriptional cofactors, the LexA yeast two-hybrid system was used. cDNAs of the ligand-binding domains of wild-type TR and TR2spl were subcloned into LexA expression vector, and coactivators, SRC-1 and transcription intermediary factor 2 (TIF2), and corepressors (NCoR and SMRT) were subcloned into B42AD expression vector. As expected, the ligand-binding domain of wild-type TR interacted with both SRC-1 and TIF2 in the presence of ligand and interacted with both NCoR and SMRT in the absence of ligand (Table 1). In contrast, the ligand-binding domain of TR2spl did not interact with any of these cofactors either in the presence or absence of ligand. Ando et al. • Aberrant Splicing of TR mRNA in a TSHoma Mol Endocrinol, September 2001, 15(9):1529–1538 1533 Fig. 4. Analysis of Splice Sites A, Splice sites used for TR2spl (red) and normal splice sites (blue and black) are depicted. B, 3⬘-ss, 5⬘-ss, and branch site of TR2spl were compared with naturally occurring splice sequences flanking exon 6 of TR2, and consensus sequences. The branch site of the deletion of TR2spl mRNA is located 48 bases upstream of the 3⬘-ss of the deletion. Underlined nucleotides are absolutely required for splicing. Capitalized nucleotides of the consensus splice sequences are important, whereas lower case nucleotides are less important. Note that TR2 exon 6 is the same as TR1 exon 9. DISCUSSION We have found a novel alternatively spliced variant of thyroid hormone receptor, TR2spl, from a TSHoma. This is also the first example of abnormal TR in TSHomas. TR2spl lacked T3-binding activity and was unable to mediate T3-dependent negative regulation of glycoprotein ␣-subunit and TSH genes. Furthermore, TR2spl showed dominant negative activity, as it blocked T3-dependent regulation of both these genes by wild-type TR2. These findings strongly implicate TR2spl in the defective negative regulation of TSH secretion exhibited by the tumor. This dysregulation in TSH secretion is similar to that observed in patients with RTH who have germline mutations in one of their TR alleles (12). Additionally, transgenic mice that overexpress dominant negative mutant TRs in the pituitary have similar defects in the negative regulation of TSH (21–23). In this connection, we recently examined TR in five other TSHomas. Although we did not find another example of aberrant alternative splicing of TRs, we identified a somatic mutation in the ligandbinding domain of TR1 in one TSHoma (24). Gittoes et al. (25) recently reported decreased expression of 1534 Mol Endocrinol, September 2001, 15(9):1529–1538 Ando et al. • Aberrant Splicing of TR mRNA in a TSHoma Fig. 5. T3-Binding Activity of the Wild-Type TR2, TR2spl, and TR2Mf Wild-type TR2, TR2spl, or TR2Mf was transfected into CV-1 cells. Nuclear extracts were prepared and incubated with 125I-T3 as described in Materials and Methods. Bound and free 125I-T3 were separated, and the ratio between the bound and total T3 (B/T) was calculated. The specific T3 binding was obtained by subtracting the nonspecific binding determined in the incubation mixture containing a 10,000fold excess of nonlabeled T3. The value of T3-binding activity of the wild-type TR2 is expressed as 100%. The data are represented as the mean ⫾ SD of four samples. TR␣ and TR in two TSHomas; therefore, downregulation of TRs may be an additional mechanism for defective negative regulation of TSH by thyroid hormone. Furthermore, unlike GH-secreting adenomas, which can harbor G protein ␣ chain (Gs␣) mutations that inhibit GTPase activity (26), no such mutations or TRH receptor mutations have been implicated in the constitutive expression of TSH in TSHomas (27). Taken together with our study, these findings suggest that defective negative regulation by the TR signaling pathway, rather than overactivity of the TRH signaling pathway, may be operative in some TSHomas. TR2spl had a 46-amino acid deletion replaced by a newly created isoleucine, which maintained the original sequence of the remaining amino acids. The deletion is located in a region of the TR ligand-binding domain where a cluster of mutations has been reported for RTH patients (12). This region also is known to contribute to the formation of helices 7, 8, and 9 of TR, which compose part of the ligand-binding pocket of TR (28). Thus, the lack of T3 binding and transcriptional activity, as well as the dominant negative activity observed for TR2spl, are fully consistent with a deletion in this important region of the TR2 ligandbinding domain. The inability of the ligand-binding domain of TR2spl to interact with corepressors and coactivators in the yeast two-hybrid system is consistent with the loss of both T3-independent activation and T3-dependent negative regulation on glycoprotein hormone ␣-subunit and TSH genes observed in our cotransfection studies. Notably, helices 3, 4, 5, and 6 of TR and helices 3, 5, 6, and 12 of TR have been reported to contact surfaces with corepressors and coactivators, respectively (29–31). Although the dele- Fig. 6. Transcriptional Activity of TR2 and TR2spl on Genes Encoding TSH Subunits Equal amounts of pcDNA expression vectors for TR2 and TR2spl were cotransfected with glycoprotein ␣-subunit or TSH luciferase reporter vectors into TSA 201 cells as described in Materials and Methods. The cells were incubated with 50 nM T3 for 40 h and harvested, and luciferase activity was measured. Data are represented as the mean ⫾ SD from nine individual samples. The asterisk indicates significant difference between TR2spl-dependent and TR2-dependent luciferase activity in the presence of T3 (P ⬍ 0.01) determined by unpaired t tests. A, Glycoprotein hormone ␣-subunit luciferase activity. B, TSH luciferase activity. tion of TR2spl is not within these areas, tertiary structure changes, presumably due to the large conformational changes caused by the deletion, are likely responsible for the loss of interaction with corepressors. Tertiary structure changes also likely affected T3-binding activity and/or coactivator interactions of TR2spl. In our study, only a small amount of wild-type TR2 mRNA was detected in the TSHoma, suggesting that TR2spl mRNA may have been spliced after wild-type TR2 mRNA was generated. Indeed, the 5⬘-ss score and 3⬘-ss score of the TR2spl ss were weaker than those of the naturally occurring ss flanking exon 6 of TR2. It is interesting that the variant form of TR1 mRNA was not observed in the TSHoma, particularly since TR1 and TR2 have identical sequences coding for the ligand-binding domain. It is possible that TR1 and TR2 mRNA have different higher order structures or differences in the turnover and/or export rates from the splicesome to account for this difference in splicing of the RNA between the two TR isoforms. Moreover, although TR2 and TR␣1 have Ando et al. • Aberrant Splicing of TR mRNA in a TSHoma Fig. 7. Dominant Negative Activity of TR2spl and TR2Mf on TR2 Regulation of Genes Encoding TSH Subunits TSA 201 cells were transfected with equal amounts of TR2 and TR2spl or TR2Mf with glycoprotein hormone ␣-subunit or TSH luciferase vector. The cells were incubated with 50 nM T3 and harvested, and luciferase activity was measured. Fold T3 inhibition is calculated as luciferase activity in the absence of T3 divided by luciferase activity in the presence of T3. Data are represented as the mean ⫾ SD from nine individual samples. A, Glycoprotein hormone ␣-subunit luciferase activity. B, TSH luciferase activity. high homology in the mRNA sequences encoding their respective ligand-binding domains, it is unlikely that TR␣1 has a corresponding splice variant, since TR␣1 has GC/AG instead of GT/AG in the critical sequences that flank the splice junction of TR2spl. TR␣ mRNA can undergo alternative splicing to generate mRNA for a non-T3 binding protein, c-erbA␣-2 (3). However, the location of the ss of TR2spl mRNA is more upstream than the 5⬘-ss of c-erbA␣-2 mRNA; therefore, the generation of TR2spl mRNA is distinct from this naturally occurring process. There are several potential mechanisms by which abnormal alternative splicing could generate TR2spl mRNA (32, 33). One possibility is the creation of a new cryptic splice site by a mutation(s) in the genomic DNA. However, this was not observed, as genomic DNA from the patient’s TSHoma did not have any mutations in the sequences flanking the splice junction or within the spliced sequence. Another possibility is that a mutation(s) in the natural ss of the flanking introns might create a less favorable splicing site, and thus allow splicing via the intraexonic ss. However, the intraexonic splicing pattern, which maintains portions of exon 6, and the absence of mutations in the naturally occurring ss contained in the intronic sequences immediately flanking exon 6, argue against this possibility. Thus, the most likely explanation for the aberrant splicing is increased activity of splicing factors in the TSHoma. The basis for this phenomenon could be Mol Endocrinol, September 2001, 15(9):1529–1538 1535 a mutation in a splicing factor(s), or increased splicing activity as a consequence of overexpression or altered function (perhaps by phosphorylation or other regulatory events) of splicing factor(s) (34). Of note, aberrant alternative splicing and overactivity of splicing factors has been recently reported in breast cancer (35–38). Additionally, aberrant splicing has been reported for ER mRNA in breast cancer (39, 40). Given the aberrant splicing of TR2 mRNA in the TSHoma, it is possible that other mRNAs may be aberrantly spliced in the tumor and perhaps contribute to the growth of the tumor. Recently, an incidental pituitary adenoma in a patient with RTH was reported (41). In addition, there is one case report of pituitary enlargement in a patient with RTH in whom pituitary regression occurred after thyroid hormone treatment (42). However, TR knockout mice and transgenic mice overexpressing mutant TR showed defective TSH regulation but no increase in tendency to form TSHomas (11, 21, 22, 43). Additionally, there have been no reported cases of TSHomas in RTH patients who have TR mutations (12, 14), and patients with RTH do not seem to have a higher risk for TSHomas. Mutant TRs per se may not be sufficient for the abnormal growth characteristics of TSHomas. Hence, the defective TR splicing could be a consequence rather than a cause of tumorigenesis. Several families with RTH have been described who do not have mutations in their TR␣ and TR genes (44). Although knockout mice that lack a coactivator have mild central thyroid hormone resistance (45), no such postreceptor defects have been detected in these patients. Thus, it is possible that aberrantly spliced TRs may account for thyroid hormone resistance in some of these families. Hormone resistance in humans has been described for estrogen, glucocorticoid, androgen, and vitamin D, as well as peptide hormones, such as insulin, vasopressin, and PTH (46– 52). In many cases, mutations in the nuclear hormone receptors or peptide hormone receptors were detected in genomic DNA and accounted for the clinical picture of hormone resistance. However, in rare instances in which no receptor mutation was found, receptor down-regulation or postreceptor defects have been described or hypothesized (44, 51, 53, 54). Our findings suggest that aberrant alternative splicing is another mechanism for hormone resistance in signaling pathways in which no receptor mutation is found in the genomic DNA. In conclusion, we describe a splicing variant of TR from a TSH-secreting pituitary tumor that caused defective T3-dependent negative regulation of glycoprotein hormone ␣-subunit and TSH genes in a transient transfection system. Since the TR gene did not contain any mutations that accounted for the deletion, this abnormal TR was caused by aberrant alternative splicing of TR2 mRNA. This example enlarges our understanding of the possible mechanisms for hormone resistance and suggests that posttranscriptional causes need to be considered whenever genomic mutations are not detected in hormone receptors. 1536 Mol Endocrinol, September 2001, 15(9):1529–1538 Ando et al. • Aberrant Splicing of TR mRNA in a TSHoma Table 1. Interactions between TR and Cofactors in a Yeast Two-Hybrid System B42AD B42AD-SRC1 B42AD-TIF2 B42AD-SMRT B42AD-NCoR LexA ⫺T3 ⫹T3 ⫺ ⫺ ⫺ ⫺ ⫺ ⫺ ⫺ ⫺ ⫺ ⫺ LexA-TRLBD ⫺T3 ⫹T3 ⫺ ⫺ ⫺ ⫹ ⫺ ⫹ ⫹ ⫺ ⫹ ⫺ LexA-TR2splLBD ⫺T3 ⫹T3 ⫺ ⫺ ⫺ ⫺ ⫺ ⫺ ⫺ ⫺ ⫺ ⫺ The expression plasmids producing either LexA DNA-binding-domain fusion proteins (listed at left) or pB42AD that encode B42 transactivation-domain fusion proteins (listed at top) were introduced into EGY48 cells, and the resulting transformants were grown on plates lacking leucine and containing X-Gal in the presence (⫹T3) or absence (⫺T3) of T3 (1 M). Two-hybrid plasmids that resulted in production of blue colored colonies are indicated as (⫹). MATERIALS AND METHODS Analyses of RNA and Genomic DNA Genomic DNA was isolated from the patient’s TSHoma and peripheral blood leukocytes. Total RNA was extracted from the TSHoma by Trizol reagent (Life Technologies, Inc., Gaithersburg, MD). Pituitary gland polyA RNA (pool of 87 normal tissue specimens) was purchased from CLONTECH Laboratories, Inc. (Palo Alto, CA). cDNA was synthesized by Moloney murine leukemia virus-reverse transcriptase. DNA was amplified by PCR with the following specific oligonucleotide primers: TR1 sense primer: 5⬘-GGATCCAGAATGATTACTAACCTATGACTC-3⬘; TR2 sense primer: 5⬘-ACCAGGGAAACAAAATGAACTACTGTATGC-3⬘; TR1 and TR2 common antisense primer: 5⬘-GGAATTATAGGAAGGAATCCAGTCAGTCTA-3⬘; TR2 exon 6 sense primer (corresponding to exon 9 of TR1): 5⬘-CTGCCATGTGAAGACCAGATCATCCT-3⬘; TR2 exon 6 antisense primer: 5⬘CTGAAGACATCAGCAGGACGGCCTGA-3⬘; Sense primer sequence for TR2 exon 6 and flanking introns: 5⬘-TCACAGAAGGTTATTCCTATT-3⬘; antisense primer sequence for TR2 exon 6 and flanking introns: 5⬘-ACTCAAGTGATTGGAATTAG-3⬘; GAPDH sense primer: 5⬘-CATCACGCCACAGTTTCCCGGAGG-3⬘; GAPDH antisense primer: 5⬘-TTTCTAGACGGCAGGTCAGGTCCACC-3⬘. For semiquantitative RTPCR, PCR conditions for temperature were optimized for each primer pair, and comparative kinetic analyses were performed to determine that the selected PCR cycle number for each primer pair was within the exponential phase of product generation. PCR products were visualized on 1% Tris-borate-EDTA agarose gels stained with ethidium bromide. Ethidium bromide-stained gels were digitized, and densitometry was performed on the product bands using Image Quant (Molecular Dynamics, Inc., Sunnyvale, CA) (55, 56). Quantification of RT-PCR by analysis of ethidium-stained gels yielded consistent results. PCR products were subcloned into pCR4TOPO plasmid (Invitrogen, Carlsbad, CA), and then sequenced. ss Profile Analysis ss Profiles were analyzed by Michael Zhang’s Exon Finder, a software program designed to predict ss sequences (19). Construction of TR cDNA Expression Vectors The full-length human wild-type TR2 cDNA was cloned into pcDNA. The DNA fragment carrying the deletion mutation identified in the patient’s TSHoma was digested with Tth111I and BglII and then subcloned into the corresponding TR2 region. A natural mutant TR1G345R from a patient with RTH was also digested with Tth111I and BglII and replaced with the corresponding TR2 region to create TR2G360R (TR2Mf). The final constructs were verified by sequencing. T3 Binding Assay T3 binding affinity of wild-type TR2 and TR2spl protein was measured in transfected CV-1 cells as described previously (57). CV-1 cells were transiently transfected with wild-type TR2, TR2spl, or TR2Mf using Lipofectamine Plus (Life Technologies, Inc.). Cells were homogenized in 0.25 M sucrose and 3 mM MgCl2 (SM). The crude nuclear pellet was suspended once with and once without 0.5% Triton X-100 in SM. Nuclear proteins were extracted by stirring isolated nuclei at 4 C for 1 h in 0.4 M KCl, 5 mM MgCl2, 1 mM dithiothreitol, and 0.05 M Tris-glycine HCl (pH 8.5). After nuclear extracts were centrifuged at 117,000 ⫻ g for 45 min, T3-binding activities in the supernatants were assayed. Nuclear proteins (125 g) were incubated with 1 nM 125I-T3 (NEN Life Science Products, Boston, MA) and 4 nM non-labeled T3 at 4 C overnight. 125I-T3 was separated from the bound form using AG1-X8 resin (Bio-Rad Laboratories, Inc., Richmond, CA). Nonspecific binding was determined in the incubation mixture containing a 10,000-fold amount of nonlabeled T3. Cotransfection Studies of TR TSA 201 cells (a strain of HEK293 cells transformed with T antigen) were maintained in Opti-MEM (Life Technologies, Inc.) containing 4% FCS pretreated with AG1-X8 resin to remove endogenous thyroid hormones. The cells were plated in six-well dishes and transfected 24 h later by the calcium phosphate precipitation method with 250 ng of TR expression vector and 500 ng of human glycoprotein hormone ␣-subunit promoter (⫺846 to ⫹44) linked to pA3 luciferase reporter gene (6). Additionally, TSA201 cells were transfected with 400 ng of TR expression vector, 200 ng of human TSH promoter (⫺1,192 to ⫹37) gene linked to pA3 luciferase reporter gene (15), and 200 ng of human thyrotroph embryonic factor expression vector (obtained by RT-PCR) with Lipofectamine Plus. Eight hours after transfection, the cells were washed and incubated for 40 h with Opti-MEM media containing 1% of resin-treated FCS with and without 50 nM T3. The cells were lysed and assayed for luciferase activity. Luciferase activity was normalized according to protein concentration. Yeast Two-Hybrid System EGY48 yeast cells, p8op-lacZ, and LexA-parental vectors (pGilda) and B42-parental vectors (pB42AD) were purchased Ando et al. • Aberrant Splicing of TR mRNA in a TSHoma from CLONTECH Laboratories, Inc. Fragments of human wild-type TR2 and TR2spl ligand-binding domain (amino acids 189–476) were subcloned into EcoRI and SalI sites of pGilda to make LexA-TRLBD and LexATRb2spl LBD. Fragments of human F-SRC-1 (1–1440), human TIF2 (1–1464), human NCoR (982–1495), and human SMRT (982–1495) were subcloned into pB42AD to make B42AD-SRC-1, B42AD-TIF2, B42AD-NCoR, and B42AD-SMRT. LexA fusion vectors, B42AD fusion vectors, and p8op-lacZ were transformed into EGY48 according to the manufacturer’s manual. Interactions of fusion proteins were tested using selection for leucine auxotrophy and Lac Z reporter gene on plates in the presence and absence of 1 M T3. Acknowledgments We thank Dr. Frederic Wondisford (University of Chicago, Chicago, IL) for human TR2 expression and TSH reporter vectors, Dr. Laird Madison, Dr. Larry Jameson, Mr. Kevin Long (Northwestern University, Chicago, IL) for glycoprotein hormone ␣-subunit reporter vector and TSA 201 cells, Dr. Ronald Evans (Salk Institute, La Jolla, CA) for human SMRT expression vector, Dr. Anthony Hollenberg (Beth Israel Hospital, Boston, MA) for human NCoR (NCoRi) expression vector, and Dr. Pierre Chambon (INSERM, Strasbourg, France) for TIF2 expression vector. We also thank Drs. Thomas Misteli and Carl Baker (National Cancer Institute, Bethesda, MD) for helpful discussions on splicing mechanisms. Received February 12, 2001. Accepted May 23, 2001. Address requests for reprints to: Paul M. Yen, M.D. Molecular Regulation and Neuroendocrinology Section, Clinical Endocrinology Branch, NIDDK, National Institutes of Health, Bethesda, Maryland 20892. E-mail: [email protected]. nih.gov. This work was supported in part by NIH Grants HG-01696 (to M.Q.Z.) and DK-15070 (to S.R.). REFERENCES 1. Brucker-Davis F, Oldfield EH, Skarulis MC, Doppman JL, Weintraub BD 1999 Thyrotropin-secreting pituitary tumors: diagnostic criteria, thyroid hormone sensitivity, and treatment outcome in 25 patients followed at the National Institutes of Health. J Clin Endocrinol Metab 84:476–486 2. Beck-Peccoz P, Brucker-Davis F, Persani L, Smallridge RC, Weintraub BD 1996 Thyrotropin-secreting pituitary tumors. Endocr Rev 17:610–638 3. Lazar MA 1993 Thyroid hormone receptors: multiple forms, multiple possibilities. Endocr Rev 14:348–399 4. Brent GA 1994 The molecular basis of thyroid hormone action. N Engl J Med 331:847–853 5. McKenna NJ, Lanz RB, O’Malley BW 1999 Nuclear receptor coregulators: cellular and molecular biology. Endocr Rev 20:321–344 6. Tagami T, Madison LD, Nagaya T, Jameson JL 1997 Nuclear receptor corepressors activate rather than suppress basal transcription of genes that are negatively regulated by thyroid hormone. Mol Cell Biol 17: 2642–2648 7. Tagami T, Park Y, Jameson JL 1999 Mechanisms that mediate negative regulation of the thyroid-stimulating hormone ␣ gene by the thyroid hormone receptor. J Biol Chem 274:22345–22353 8. Wood WM, Dowding JM, Haugen BR, Bright TM, Gordon DF, Ridgway EC 1994 Structural and functional charac- Mol Endocrinol, September 2001, 15(9):1529–1538 1537 terization of the genomic locus encoding the murine 2 thyroid hormone receptor. Mol Endocrinol 8:1605–1617 9. Hodin RA, Lazar MA, Chin WW 1990 Differential and tissue-specific regulation of the multiple rat c-erbA mRNA species by thyroid hormone. J Clin Invest 85: 101–105 10. Oppenheimer JH, Schwartz HL 1997 Molecular basis of thyroid hormone-dependent brain development. Endocr Rev 18:462–475 11. Abel ED, Boers ME, Pazos-Moura C, et al. F 1999 Divergent roles for thyroid hormone receptor  isoforms in the endocrine axis and auditory system. J Clin Invest 104: 291–300 12. Refetoff S, Weiss RA, Usala SJ 1993 The syndromes of resistance to thyroid hormone. Endocr Rev 14:348–399 13. Chatterjee VK, Beck-Peccoz P 1994 Hormone receptor interactions in health and disease. Thyroid hormone resistance. Balliere’s Clin Endocrinol Metab 8:267–283 14. Brucker-Davis F, Skarulis MC, Grace MB, et al. 1995 Genetic and clinical features of 42 kindreds with resistance to thyroid hormone. The National Institutes of Health Prospective Study. Ann Intern Med 123:572–583 15. Bodenner DL, Mroczynski MA, Weintraub BD, Radovick S, Wondisford FE 1991 A detailed functional and structural analysis of a major thyroid hormone inhibitory element in the human thyrotropin -subunit gene. J Biol Chem 266:21666–21673 16. Tagami T, Jameson JL 1998 Nuclear corepressors enhance the dominant negative activity of mutant receptors that cause resistance to thyroid hormone. Endocrinology 139:640–650 17. Shapiro MB, Senapathy P 1987 RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res 15:7155–7174 18. Zhang MQ 1998 Statistical features of human exons and their flanking regions. Hum Mol Genet 7:919–932 19. Zhang MQ 1997 Identification of protein coding regions in the human genome by quadratic discriminant analysis. Proc Natl Acad Sci USA 94:565–568 20. Weiss RE, Refetoff S 2000 Resistance to thyroid hormone. Rev Endocr Metab Disord 1:97–108 20a.Sakurai A, Miyamoto T, Refetoff S, DeGroot LJ 1990 Dominant negative transcriptional regulation by a mutant thyroid hormone receptor-beta in a family with generalized resistance to thyroid hormone. Mol Endocrinol 4:1988–1994 21. Kaneshige M, Kaneshige K, Zhu X, et al. 2000 Mice with a targeted mutation in the thyroid hormone  receptor gene exhibit impaired growth and resistance to thyroid hormone. Proc Natl Acad Sci USA 97:13209–13214 22. Abel ED, Kaulbach HC, Campos-Barros A, et al. 1999 Novel insight from transgenic mice into thyroid hormone resistance and the regulation of thyrotropin. J Clin Invest 103:271–279 23. Hayashi Y, Xie J, Weiss RE, Pohlenz J, Refetoff S 1998 Selective pituitary resistance to thyroid hormone produced by expression of a mutant thyroid hormone receptor  gene in the pituitary gland of transgenic mice. Biochem Biophys Res Commun 245:204–210 24. Ando S, Sarlis NJ, Krishnan J, Feng X, Oldfield E, Yen PM 2001 Somatic mutation or aberrant alternative-splicing of thyroid hormone receptor beta can cause a defect in negative-regulation of thyrotropin in thyrotropin-secreting pituitary tumors. Proc 83rd Annual Meeting of The Endocrine Society, Denver, CO, 2001, p 98 (Abstract OR23-3) 25. Gittoes NJ, McCabe CJ, Verhaeg J, Sheppard MC, Franklyn JA 1998 An abnormality of thyroid hormone receptor expression may explain abnormal thyrotropin production in thyrotropin-secreting pituitary tumors. Thyroid 8:9–14 1538 Mol Endocrinol, September 2001, 15(9):1529–1538 26. Landis CA, Masters SB, Spada A, Pace AM, Bourne HR, Vallar L 1989 GTPase inhibiting mutations activate the alpha chain of Gs and stimulate adenylyl cyclase in human pituitary tumours. Nature 340:692–696 27. Dong Q, Brucker-Davis F, Weintraub BD, et al. 1996 Screening of candidate oncogenes in human thyrotroph tumors: absence of activating mutations of the G ␣ q, G ␣ 11, G ␣ s, or thyrotropin-releasing hormone receptor genes. J Clin Endocrinol Metab 81:1134–1140 28. Wagner RL, Apriletti JW, McGrath ME, West BL, Baxter JD, Fletterick RJ 1995 A structural role for hormone in the thyroid hormone receptor. Nature 378:690–697 29. Perissi V, Staszewski LM, McInerney EM, et al. 1999 Molecular determinants of nuclear receptor-corepressor interaction. Genes Dev 13:3198–3208 30. Nagy L, Kao HY, Love JD, et al. 1999 Mechanism of corepressor binding and release from nuclear hormone receptors. Genes Dev 13:3209–3216 31. Feng W, Ribeiro RC, Wagner RL, et al. 1998 Hormonedependent coactivator binding to a hydrophobic cleft on nuclear receptors. Science 280:1747–1749 32. Minvielle-Sebastia L, Keller W 1999 mRNA polyadenylation and its coupling to other RNA processing reactions and to transcription. Curr Opin Cell Biol 11:352–357 33. Philips AV, Cooper TA 2000 RNA processing and human disease. Cell Mol Life Sci 57:235–249 34. Misteli T 1999 RNA splicing: what has phosphorylation got to do with it? Curr Biol 9:R198–200 35. Lee MP, Feinberg AP 1997 Aberrant splicing but not mutations of TSG101 in human breast cancer. Cancer Res 57:3131–3134 36. Zhu X, Daffada AA, Chan CM, Dowsett M 1997 Identification of an exon 3 deletion splice variant androgen receptor mRNA in human breast cancer. Int J Cancer 72:574–580 37. Silberstein GB, Van Horn K, Strickland P, Roberts Jr CT, Daniel CW 1997 Altered expression of the WT1 Wilms tumor suppressor gene in human breast cancer. Proc Natl Acad Sci USA 94:8132–8137 38. Stickeler E, Kittrell F, Medina D, Berget SM 1999 Stagespecific changes in SR splicing factors and alternative splicing in mammary tumorigenesis. Oncogene 18: 3574–3582 39. McGuire WL, Chamness GC, Fuqua SA 1991 Estrogen receptor variants in clinical breast cancer. Mol Endocrinol 5:1571–1577 40. Iwase H, Omoto Y, Iwata H, Hara Y, Ando Y, Kobayashi S 1998 Genetic and epigenetic alterations of the estrogen receptor gene and hormone independence in human breast cancer. Oncology 1:11–16 41. Safer JD, Colan SD, Fraser LM, Wondisford FE 2001 A pituitary tumor in a patient with thyroid hormone resistance: a diagnostic dilemma. Thyroid 11:281–291 42. Gurnell M, Rajanayagam O, Barbar I, Jones MK, Chatterjee VK 1998 Reversible pituitary enlargement in the Ando et al. • Aberrant Splicing of TR mRNA in a TSHoma 43. 44. 45. 46. 47. 48. 49. 50. 51. 52. 53. 54. 55. 56. 57. syndrome of resistance to thyroid hormone. Thyroid 8:679–682 Gothe S, Wang Z, Ng L, et al. 1999 Mice devoid of all known thyroid hormone receptors are viable but exhibit disorders of the pituitary-thyroid axis, growth, and bone maturation. Genes Dev 13:1329–1341 Pohlenz J, Weiss RE, Macchia PE, et al. 1999 Five new families with resistance to thyroid hormone not caused by mutations in the thyroid hormone receptor  gene. J Clin Endocrinol Metab 84:3919–3928 Weiss RE, Xu J, Ning G, Pohlenz J, O’Malley BW, Refetoff S 1999 Mice deficient in the steroid receptor coactivator 1 (SRC-1) are resistant to thyroid hormone. EMBO J 18:1900–1904 Smith EP, Boyd J, Frank GR, et al. 1994 Estrogen resistance caused by a mutation in the estrogen-receptor gene in a man. N Engl J Med 331:1056–1061 Chrousos GP, Detera-Wadleigh SD, Karl M 1993 Syndromes of glucocorticoid resistance. Ann Intern Med 119:1113–1124 Malloy PJ, Pike JW, Feldman D 1999 The vitamin D receptor and the syndrome of hereditary 1,25-dihydroxyvitamin D-resistant rickets. Endocr Rev 20:156–188 Patterson MN, McPhaul MJ, Hughes IA 1994 Androgen insensitivity syndrome. Baillieres Clin Endocrinol Metab 8:379–404 Bichet DG 1998 Nephrogenic diabetes insipidus. Am J Med 105:431–442 Spiegel AM 1996 Defects in G protein-coupled signal transduction in human disease. Annu Rev Physiol 58: 143–170 Krook A, O’Rahilly S 1996 Mutant insulin receptors in syndromes of insulin resistance. Baillieres Clin Endocrinol Metab 10:97–122 Lazar MA 1999 Nuclear hormone receptors: from molecules to diseases. J Invest Med 47:364–368 Chrousos GP 1999 A new “new” syndrome in the new world: is multiple postreceptor steroid hormone resistance due to a coregulator defect? J Clin Endocrinol Metab 84:4450–4453 Nakayama H, Yokoi H, Fujita J 1992 Quantification of mRNA by non-radioactive RT-PCR and CCD imaging system. Nucleic Acids Res 20:4939 Gittoes NJ, McCabe CJ, Verhaeg J, Sheppard MC, Franklyn JA 1997 Thyroid hormone and estrogen receptor expression in normal pituitary and nonfunctioning tumors of the anterior pituitary. J Clin Endocrinol Metab 82:1960–1967 Ando S, Nakamura H, Sasaki S, et al. 1996 Introducing a point mutation identified in a patient with pituitary resistance to thyroid hormone (Arg 338 to Trp) into other mutant thyroid hormone receptors weakens their dominant negative activities. J Endocrinol 151:293–300