* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Overview of genome analysis Page 421
Survey
Document related concepts
Transcript
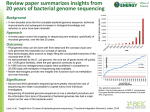
Completed Genomes: Viruses and Bacteria Monday, October 20, 2003 Introduction to Bioinformatics ME:440.714 J. Pevsner [email protected] Copyright notice Many of the images in this powerpoint presentation are from Bioinformatics and Functional Genomics by J Pevsner (ISBN 0-471-21004-8). Copyright © 2003 by Wiley. These images and materials may not be used without permission from the publisher. Visit http://www.bioinfbook.org Announcements We are now beginning the last third of the course: Today: completed genomes (Chapters 12-14) Wednesday: Fungi. Exam #2 is due at the start of class. Next Monday: Functional genomics (Jef Boeke) Next Wednesday: Pathways (Joel Bader) Monday Nov. 3: Eukaryotic genomes Wednesday Nov. 5: Human genome Monday Nov. 10: Human disease Wednesday Nov. 12: Final exam (in class) Outline of today’s lecture Genome projects (Chapter 12) chronological overview major issues and themes Introduction to viruses (Chapter 13) classification bioinformatics challenges and resources Introduction to bacteria and archaea (Chapter 14) classification bioinformatics challenges and resources Introduction to genomes A genome is the collection of DNA that comprises an organism. Today we have assembled the sequence of hundreds of genomes. We will begin by introducing the “tree of life” in an effort to make a comprehensive survey of life forms. Page 397 Introduction: Systematics Ernst Haeckel (1834-1919), a supporter of Darwin, published a tree of life (1879) including Moner (formless clumps, later named bacteria). Chatton (1937) distinguished prokaryotes (bacteria that lack nuclei) from eukaryotes (having nuclei). Whittaker and others described the five-kingdom system: animals, plants, protists, fungi, and monera. In the 1970s and 1980s, Carl Woese and colleagues described the archaea, thus forming a tree of life with three main branches. Page 399 Five kingdom system (Haeckel, 1879) mammals vertebrates animals invertebrates plants fungi protists monera protozoa Page 396 Pace (2001) described a tree of life based on small subunit rRNA sequences. This tree shows the main three branches described by Woese and colleagues. Fig. 12.1 Page 400 Molecular sequences as basis of trees Historically, trees were generated primarily using characters provided by morphological data. Molecular sequence data are now commonly used, including sequences (such as small-subunit RNAs) that are highly conserved. Visit the European Small Subunit Ribosomal RNA database for 20,000 SSU rRNA sequences. Page 401 Genome sequencing projects Genomes that span the tree of life are being sequenced at a rapid rate. There are several web-based resources that document the progress, including: GNN Genome News Network http://www.genomenewsnetwork.org/main.shtml GOLD Genomes Online Database http://wit.integratedgenomics.com/GOLD/ PEDANT Protein Extraction, Description & Analysis Tool http://pedant.gsf.de/ Page 405 Genome sequencing projects There are three main resources for genomes: EBI European Bioinformatics Institute http://www.ebi.ac.uk/genomes/ NCBI National Center for Biotechnology Information http://www.ncbi.nlm.nih.gov TIGR The Institute for Genomic Research http://www.tigr.org Page 405 archaea bacteria eukaryota http://www.ncbi.nlm.nih.gov/Entrez/ Overview of viral complete genomes Overview of archaea complete genomes Overview of eukaryota genomes in NCBI’s Entez division Overview of eukaryota genomes in NCBI’s Entrez division Chronology of genome sequencing projects We will next summarize the major achievements in genome sequencing projects from a chronological perspective. Page 404 Chronology of genome sequencing projects 1977: first viral genome Sanger et al. sequence bacteriophage fX174. This virus is 5386 base pairs (encoding 11 genes). See accession J02482. 1981 Human mitochondrial genome 16,500 base pairs (encodes 13 proteins, 2 rRNA, 22 tRNA) Today, over 400 mitochondrial genomes sequenced 1986 Chloroplast genome 156,000 base pairs (most are 120 kb to 200 kb) Page 406 Entrez nucleotide record for bacteriophage fX174 (graphics display) Fig. 12.6 Page 407 mitochondrion chloroplast Lack mitochondria (?) Chronology of genome sequencing projects 1995: first genome of a free-living organism, the bacterium Haemophilus influenzae Page 409 1995: genome of the bacterium Haemophilus influenzae is sequenced Fig. 12.9 Page 411 Overview of bacterial complete genomes You can find functional annotation through the COGs database (Clusters of Orthologous Genes) Fig. 12.9 Page 411 Click the circle to access the genome sequence Fig. 12.9 Page 411 Genes are color-coded according to the COGs scheme Click the circle to access the genome sequence Fig. 12.10 Page 412 Chronology of genome sequencing projects 1996: first eukaryotic genome The complete genome sequence of the budding yeast Saccharomyces cerevisiae was reported. We will describe this genome on Wednesday. Also in 1996, TIGR reported the sequence of the first archaeal genome, Methanococcus jannaschii. Page 413 1996: a yeast genome is sequenced To place the sequencing of the yeast genome in context, these are the eukaryotes… Eukaryotes (Baldauf et al. 2000) Fungi Chronology of genome sequencing projects 1997: More bacteria and archaea Escherichia coli 4.6 megabases, 4200 proteins (38% of unknown function) 1998: first multicellular organism Nematode Caenorhabditis elegans 97 Mb; 19,000 genes. 1999: first human chromosome Chromosome 22 (49 Mb, 673 genes) Page 413 1999: Human chromosome 22 sequenced 1999: Human chromosome 22 sequenced 49 MB 673 genes Chronology of genome sequencing projects 2000: Fruitfly Drosophila melanogaster (13,000 genes) Plant Arabidopsis thaliana Human chromosome 21 2001: draft sequence of the human genome (public consortium and Celera Genomics) Page 415 2000 Completed genome projects (current) Eukaryotes: 10 In progress (partial): Anopheles gambiae Danio rerio (zebrafish) Arabidopsis thaliana Glycine max (soybean) Caenorhabditis elegans Hordeum vulgare (barley) Drosophila melanogaster Leishmania major Encephalitozoon cuniculi Rattus norvegicus Guillardia theta nucleomorph Mus musculus Plasmodium falciparum Saccharomyces cerevisiae (yeast) Schizosaccharomyces pombe Viruses: 1419 Bacteria: 139 Archaea: 36 Page 417 eukaryotes Overview of genome analysis [1] Selection of genomes for sequencing [2] Sequence one individual genome, or several? [3] How big are genomes? [4] Genome sequencing centers [5] Sequencing genomes: strategies [6] When has a genome been fully sequenced? [7] Repository for genome sequence data [8] Genome annotation Page 418 Fig. 12.11 Page 418 Overview of genome analysis [1] Selection of genomes for sequencing is based on criteria such as: • genome size (some plants are >>>human genome) • cost • relevance to human disease (or other disease) • relevance to basic biological questions • relevance to agriculture Page 419 Overview of genome analysis [1] Selection of genomes for sequencing is based on criteria such as: • genome size (some plants are >>>human genome) • cost • relevance to human disease (or other disease) • relevance to basic biological questions • relevance to agriculture Ongoing projects: Chicken Chimpanzee Cow Dog (recent publication) Fungi (many) Honey bee Sea urchin Rhesus macaque Page 419 Overview of genome analysis [2] Sequence one individual genome, or several? Try one… --Each genome center may study one chromosome from an organism --It is necessary to measure polymorphisms (e.g. SNPs) in large populations (November 5) For viruses, thousands of isolates may be sequenced. For the human genome, cost is the impediment. Page 419 Overview of genome analysis [3] How big are genomes? Viral genomes: 1 kb to 350 kb (Mimivirus: 800 kb) Bacterial genomes: 0.5 Mb to 13 Mb Eukaryotic genomes: 8 Mb to 686 Mb (discussed further on Monday, November 3) Page 420 Genome sizes in nucleotide base pairs plasmids viruses bacteria fungi plants algae insects mollusks bony fish The size of the human genome is ~ 3 X 109 bp; almost all of its complexity is in single-copy DNA. amphibians reptiles birds The human genome is thought to contain ~30,000-40,000 genes. 104 105 106 107 mammals 108 109 1010 1011 http://www3.kumc.edu/jcalvet/PowerPoint/bioc801b.ppt Overview of genome analysis [4] 20 Genome sequencing centers contributed to the public sequencing of the human genome. Many of these are listed at the Entrez genomes site. (See Table 17.6, page 625.) Page 421 Overview of genome analysis [5] There are two main stragies for sequencing genomes Whole Genome Shotgun (from the NCBI website) An approach used to decode an organism's genome by shredding it into smaller fragments of DNA which can be sequenced individually. The sequences of these fragments are then ordered, based on overlaps in the genetic code, and finally reassembled into the complete sequence. The 'whole genome shotgun' (WGS) method is applied to the entire genome all at once, while the 'hierarchical shotgun' method is applied to large, overlapping DNA fragments of known location in the genome. Page 421 Overview of genome analysis Hierarchical shotgun method Assemble contigs from various chromosomes, then sequence and assemble them. A contig is a set of overlapping clones or sequences from which a sequence can be obtained. The sequence may be draft or finished. A contig is thus a chromosome map showing the locations of those regions of a chromosome where contiguous DNA segments overlap. Contig maps are important because they provide the ability to study a complete, and often large segment of the genome by examining a series of overlapping clones which then provide an unbroken succession of information about that region. Page 421 Overview of genome analysis [6] When has a genome been fully sequenced? A typical goal is to obtain five to ten-fold coverage. Finished sequence: a clone insert is contiguously sequenced with high quality standard of error rate 0.01%. There are usually no gaps in the sequence. Draft sequence: clone sequences may contain several regions separated by gaps. The true order and orientation of the pieces may not be known. Page 422 Overview of genome analysis [7] Repository for genome sequence data Raw data from many genome sequencing projects are stored at the trace archive at NCBI or EBI (main NCBI page, bottom right) Page 425 Fig. 12.14 Page 426 Fig. 12.14 Page 426 Overview of genome analysis [8] Genome annotation Information content in genomic DNA includes: -- repetitive DNA elements -- nucleotide composition (GC content) -- protein-coding genes, other genes These topics will be discussed in detail on November 3 (eukaryotic genomes) Page 425 GC content varies across genomes Bacteria Number of species in each GC class 10 5 Plants 5 Invertebrates 3 Vertebrates 10 5 20 30 40 50 60 70 GC content (%) 80 Fig. 12.16 Page 428 Introduction to viruses Viruses are small, infectious, obligate intracellular parasites. They depend on host cells to replicate. Because they lack the resources for independent existence, they exist on the borderline of the definition of life. The virion (virus particle) consists of a nucleic acid genome surrounded by coat proteins (capsid) that may be enveloped in a host-derived lipid bilayer. Viral genomes consist of either RNA or DNA. They may be single-, double, or partially double stranded. The genomes may be circular, linear, or segmented. Page 437 Introduction to viruses Viruses have been classified by several criteria: -- based on morphology (e.g. by electron microscopy) -- by type of nucleic acid in the genome -- by size (rubella is about 2 kb; HIV-1 about 9 kb; poxviruses are several hundred kb). Mimivirus (for Mimicking microbe) has a double-stranded circular genome of 800 kb. -- based on human disease Page 438 Fig. 13.1 Page 439 The International Committee on Taxonomy of Viruses (ICTV) offers a website, accessible via NCBI’s Entrez site http://www.ncbi.nlm.nih.gov/ICTVdb/ Fig. 13.2 Page 440 Introduction to viruses Vaccine-preventable viral diseases include: Hepatitis A Hepatitis B Influenza Measles Mumps Poliomyelitis Rubella Smallpox Page 441 Bioinformatic approaches to viruses Some of the outstanding problems in virology include: -- Why does a virus such as HIV-1 infect one species (human) selectively? -- Why do some viruses change their natural host? In 1997 a chicken influenza virus killed six people. -- Why are some viral strains particularly deadly? -- What are the mechanisms of viral evasion of the host immune system? -- Where did viruses originate? Page 439-441 Diversity and evolution of viruses The unique nature of viruses presents special challenges to studies of their evolution. • viruses tend not to survive in historical samples • viral polymerases of RNA genomes typically lack proofreading activity • viruses undergo an extremely high rate of replication • many viral genomes are segmented; shuffling may occur • viruses may be subjected to intense selective pressures (host immune respones, antiviral therapy) • viruses invade diverse species • the diversity of viral genomes precludes us from making comprehensive phylogenetic trees of viruses Page 441 Bioinformatic approaches to herpesvirus Herpesviruses are double-stranded DNA viruses that include herpes simplex, cytomegalovirus, and Epstein-Barr. Phylogenetic analysis suggests three major groups that originated about 180-220 MYA. Page 442 Fig. 13.3 Page 443 Bioinformatic approaches to herpesvirus Consider human herpesvirus 9 (HHV-8). Its genome is about 140,000 base pairs and encodes about 80 proteins. We can explore this virus at the NCBI website. Try NCBI Entrez Genomes viruses dsDNA Page 442 Fig. 13.4 Page 444 Fig. 13.5 Page 445 Fig. 13.10 Page 449 Bioinformatic approaches to herpesvirus Consider human herpesvirus 9 (HHV-8). Its genome is about 140,000 base pairs and encodes about 80 proteins. Microarrays have been used to define changes in viral gene expression at different stages of infection (Paulose-Murphy et al., 2001). Conversely, gene expression changes have been measured in human cells following viral infection. Page 442 Paulose-Murphy et al. (2001) described HHV-8 viral genes that are expressed at different times post infection Fig. 13.11 Page 450 Bioinformatic approaches to HIV Human Immunodeficiency Virus (HIV) is the cause of AIDS. At the end of the year 2002, 42 million people were infected. HIV-1 and HIV-2 are primate lentiviruses. The HIV-1 genome is 9181 bases in length. Note that there are almost 100,000 Entrez nucleotide records for this genome (but only one RefSeq entry). Phylogenetic analyses suggest that HIV-2 appeared as a cross-species contamination from a simian virus, SIVsm (sooty mangebey). Similarly, HIV-1 appeared from simian immunodeficiency virus of the chimpanzee (SIVcpz). Page 446 Fig. 13.6 Page 446 Bioinformatic approaches to HIV Two major resources are NCBI and the Los Alamos National Laboratory (LANL) databases. See http://hiv-web.lanl.gov/ LANL offers -- an HIV BLAST server -- Synonymous/non-synonymous analysis program -- a multiple alignment program -- a PCA-like tool -- a geography tool Page 453 Fig. 13.13 Page 452 Fig. 13.6 Page 446 Bacteria and archaea: genome analysis Bacteria and archaea constitute two of the three main branches of life. Together they are the prokaryotes. We can classify prokaryotes based on six criteria: [1] morphology [2] genome size [3] lifestyle [4] relevance to human disease [5] molecular phylogeny (rRNA) [6] molecular phylogeny (other molecules) Page 466 Fig. 14.1 Page 468 M. genitalium has one of the smallest bacterial genome sizes. View its genome at www.tigr.org Fig. 14.2 Page 470 Bacteria and archaea: lifestyles We may distinguish six prokaryotic lifestyles: [1] Extracellular (e.g. E. coli) [2] Facultatively intracellular (Mycobacterium tuberculosis) [3] Extremophilic (e.g. M. jannaschi) [4] epicellular bacteria (e.g. Mycoplasma pneumoniae) [5] obligate intracellular and symbiotic (B. aphidicola) [6] obligate intracellular and parasitic (Rickettsia) Page 472 Fig. 14.4 Page 477 Revised figure Fig. 14.5 Page 478 Fig. 14.6 Page 479 DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae Nature 406, 477- 483 (2000) Bacteria and archaea: finding genes Four main features of genomic DNA are useful: [1] Open reading frame length [2] Consensus for ribosome binding (Shine-Dalgarno) [3] Pattern of codon usage [4] Homology of putative gene to other genes Page 480 GLIMMER for gene-finding in bacteria (www.tigr.org) Fig. 14.7 Page 482 Lateral gene transfer occurs in stages Fig. 14.8 Page 484 COGs database: organisms and tools COGs database: functional annotation COGs database: distribution of COGs by number of clades... COGs database: distribution of COGs by number of species How can whole genomes be compared? -- molecular phylogeny -- You can BLAST (or PSI-BLAST) all the DNA and/or protein in one genome against another -- TaxPlot and COG for bacterial (and for some eukaryotic) genomes -- PipMaker, MUMmer and other programs align large stretches of genomic DNA from multiple species Fig. 14.16 Page 493 Fig. 14.16 Page 493 Fig. 14.17 Page 494 Fig. 14.18 Page 495