* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download IGEM2006-UCSF-Powerpoint
E. coli long-term evolution experiment wikipedia , lookup
Gel electrophoresis of nucleic acids wikipedia , lookup
Non-coding DNA wikipedia , lookup
Molecular cloning wikipedia , lookup
Molecular evolution wikipedia , lookup
Biosynthesis wikipedia , lookup
DNA supercoil wikipedia , lookup
Community fingerprinting wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Transformation (genetics) wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
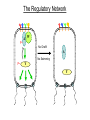
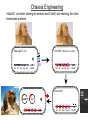
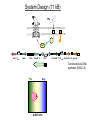
Remote Control of Bacterial Chemotaxis UCSF iGEM Team 2006 Patrick Visperas Matthew Eames Eli Groban Ala Trusina Christopher Voigt, Tanja Kortemme, Chao Tang, Chris Rao Goal Remote Control of a Bacterial Machine Start How Bacterial Chemotaxis Works Bacteria Swim Up Gradients How Chemotaxis is Observed Goulian Motility Assay (U Penn) Glide CCW CW concentration gradient No gradient ? Tumble The Regulatory Network A W P~ No CheW A No Swimming P~ Y Y Binding Partners •Orthogonal binding pair Tsr-CheW (Liu et al, 1991) •We mapped these mutations onto the Tar-Chew complex Tar CheW Tsr mutation can be mapped to Tar Gradient Aspartate • point mutation in Tar* reduces motility to approximately 40% that of wild-type Part Design • New orthogonal signaling pairs “Codon randomization” is used to reuse scaffolds without fear of recombination Chassis Engineering • Build E. coli strain lacking all sensors and CheW, but retaining the other chemotaxis proteins UU1250 (Parkinson, U Utah) Wild-type E. coli tar tsr tap trg aer cheW tar tsr tap trg aer cheW UUDcheW cheW* tar* tar tsr tap trg aer cheW Device Characterization: The Fim-Switch INPUT [ara] OUTPUT MCS Ptrc* rfp IRR IRL gfp FimE FimE IRL IRR T. Ham, et al (2006) Tested in UU1250 35 30 2 Possible Problems 25 1. The plasmid is unstable 20 15 2. The strain contains FimE/ FimB 10 5 RED + ara % bacteria - ara 0 0 0.3 1.2 5 1 2 3 4 5 6 7 8 GREEN [arabinose] (mM) System Design (11 kB) Phe CheY araC PBAD fimE T1x4 CheW r2 Ptrc* r2 CheW* T1 PlacIQ r0 phe-tar r0 asp-tar* Constructed via DNA synthesis (DNA 2.0) Phe Asp - arabinose System Design (11 kB) Asp CheY araC PBAD fimE T1x4 CheW r2 Ptrc* r2 CheW* T1 PlacIQ r0 phe-tar r0 asp-tar* Constructed via DNA synthesis (DNA 2.0) Phe Asp - arabinose Phe Asp + arabinose Results Gradient Gradient Aspartate Phenylalanine Conclusions • Orthogonal pairs are a rapid method to built signaling pathways that can operate simultaneously • Chassis developed to rapidly screen for new proteinprotein interactions • Limited by the switch performance in this chassis • Directional control could also be achieved by nonchemical inputs (light, radiation, etc)