* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Bayesian Hierarchical Model for QTLs
Genome evolution wikipedia , lookup
Population genetics wikipedia , lookup
Skewed X-inactivation wikipedia , lookup
Neocentromere wikipedia , lookup
Y chromosome wikipedia , lookup
Gene expression programming wikipedia , lookup
Designer baby wikipedia , lookup
X-inactivation wikipedia , lookup
History of genetic engineering wikipedia , lookup
Microevolution wikipedia , lookup
Public health genomics wikipedia , lookup
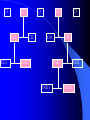
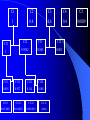
www. geocities.com/ResearchTriangle/Forum/4463/anigenetics.gif Bayesian Hierarchical Model for QTLs Susan Simmons University of North Carolina Wilmington Collaborators Dr. Edward Boone Dr. Ann Stapleton Mr. Haikun Bao DNA Chromosome Genes Genetic Map Chromosome 1 of Protozoa Cryptosporidium parvum Chromosome 1 of Homo sapiens Alleles Genetic Maps Many more maps available at www.ncbi.nih.gov Knowing information about genes now allows us to find associations between genes and outcomes (phenotypes) Some examples In 1989 a breakthrough was made for the disease of cystic fibrosis. Location (or locus) is 7q31.2 - The CFTR gene is found in region q31.2 on the long (q) arm of human chromosome 7 (single gene responsible for this disease). The disease arises when an individual has two recessive copies at this location. An individual with one dominant and one recessive is said to be a carrier of the disease. Genetic screening to determine disease. Green revolution The Green Revolution is the increase in food production stemming from the improved strains of wheat, rice, maize and other cereals in the 1960s developed by Dr Norman Borlaug in Mexico and others under the sponsorship of the Rockefeller Foundation Created new species of wheat and rice that produced higher yield. QTL Better medical treatments and increased agriculture are only two examples in which identifying the location on the genome can have an impact. Identifying the region on the genome (or on the chromosome) responsible for a quantitative trait (as opposed to qualitative as disease) is known as Quantitative Trait Locus (QTL). Existing software Zhao-Bang Zeng’s group at NC State has QTL Cartographer Karl Broman (John Hopkins) has an R program that performs a number of algorithms for QTLs To use these algorithms (and a number of other published algorithms) only one observation per genotype can be used World of plants Why plants? Increase yield to feed our increasing population Make plants resistant to UV-B exposure Plants, continued Control – Design and Environment – Reproduction – Design (RIL is one of the best designs for detecting QTLs)… Alleles are homozygous Cost Time Plant QTL experiments In most experiments, a number of replicates or clones are observed within each line A number of plant biologist use some summary measure to use conventional methods Information is lost (and can be misleading…example in Conte et al (unpublished)) Hierarchical model to incorporate replicates within each line Data Trait or phenotype, yij , i = 1,..,L where L is the number of lines and j = 1, …, ni (number of replicates within each line) Design matrix, X is L x M where M is the number of markers on the genetic map Hierarchical Model Hierarchical Model yij ~ N(li,si2) li ~ N(XiTb,t 2) Priors t 2 ~ Inverse c 2 (1) bk ~ N(0,100) si2 ~ Inverse c 2 (1) Posterior Model Probability Let denote the set of all possible models. Given data D, the posterior probability of model ki is given by Bayes Rule P ( ki | D ) P ( D | ki ) P ( ki ) P( D | k ) P(k ) j 1 i i (These probabilities are implicitly conditioned on the set ) Posterior Model continued To compute probability of the model given the ) need to data in previous slide ( P(ki),| Dwe compute P(D|ki), where P( D | ki ) P( D | qi , ki ) P(qi | ki )dqi qi is the vector of unknown parameters for model ki Integration This integration can become difficult since the length of the unknown parameters is 2*L + M +2. Use Monte Carlo estimate of the integral 1 t ( j) P ( D | q , k ) P ( q | k ) d q P ( D | q i i i i i i , ki ) t j 1 Where qi( j ) , j = 1,…,t are samples from the posterior distribution Search strategy The activation probability, P(bj 0|D) is defined as P( b j 0 | D) P( b j 0 | ki , D) P(ki | D) There are 2M number of potential models,which can make the calculation of P(bj 0|D) computationally intensive Instead, we define a conditional probability search approach C1 C2 C21 C211 C3 C22 C4 C41 C212 C5 C42 C421 C4211 C422 C4212 Simulated data Using the line information from the Bay x Sha RIL population, a single QTL was simulated on the fourth marker of the first chromosome. The Bay x Sha population has 5 chromosomes. C1 C2 C3 C4 C5 1 0.4 0.6 0.4 0.0029 C11 C12 C31 C32 1 0.9362 0.063 0.063 C111 C112 C121 C122 0.818 0.927 0.114 0.108 C1111 C1112 C1121 C1122 0.041 (M1) 0.014(M2) 0.083(M3) 1(M4) Comments Need to run model on more simulations Would like to compare this search strategy to a stochastic search Would like to include epistasis in the model Thank you