* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Document
Survey
Document related concepts
Transcript
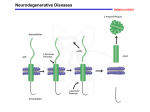
New a-Synuclein Mutants: How Do They Contribute To Parkinson’s Disease? Sara Herrera Advisor: Shubhik K. DebBurman Department of Biology Lake Forest College Road Map •Parkinson’s Disease •a-Synuclein Misfolding •Model System & Hypothesis •Results •Conclusion Neurodegeneration Disease Protein Parkinson’s Disease a-Synuclein Alzheimer’s Disease Amyloid -peptide Huntington’s Disease Huntingtin Prion Disease Prion protein Spinocerebellar Ataxia Ataxin Protein Misfolding Cell Death Parkinson’s Disease • Affects over 4 million people worldwide • Slowness of movement, resting tremors, postural instability • Death of dopaminergic neurons that control movement • Protein aggregates within these neurons Diseased Healthy Perves et al. Neuroscience, 2nd edition a-Synuclein Cytoplasmic Protein Presynaptic Terminals of neurons a-synuclein Functions Unknown 140 amino acids a-Synuclein Misfolding & Toxicity Native a-Synuclein Misfolded a-Synuclein Aggregated a-Synuclein (Lewy Bodies) Toxicity (Cell Death) Spillantini et al., 1997 Known Familial PD Mutants Wild-type a-syn Normal Gene -In all humans E46K a-syn A30P a-syn Newly Discovered, 2004 Natural Mutations -Genetic PD A53T a-syn A30P/A53T a-syn Artificial Mutation Budding Yeast Model System Why Yeast? 1. Conservation of genes 2. Sequenced Genome S. cerevisiae Prion disease model (1998) HD model (1999) PD model (2003) DebBurman Yeast Model Predictions a-syn a-syn GFP Our Model 19 kDa 28 kDa 54 kDa 62 kDa Johnson, 2003 Sharma, 2004 In our model a-synuclein runs 8-10 kDa higher on protein gels. What causes this altered migration of a-synuclein? Systematic Examination of Possible aSynuclein Modifications Post-Translational Modifications •Phosphorylation •Glycosylation •Lipidation •Ubiquitination •Nitrosylation •Oxidation Post-Translational Modification -Lee, et al. 2000, demonstrated that a-synuclein was nitrated in Lewy Bodies. -Souza, et al. 2000, demonstrated that nitrating and oxidizing agents can nitrate and oxidize a-synuclein at tyrosine residues, resulting in oligomers -Fujiwara, et al. 2003, showed that a-synuclein can be phosphorylation at Serine 129. This promotes fibril formation. Creation of Post-Translational Modification Mutants Seen in PD Patients Nitrosylation Oxidation Phosphorylation Ubiquitination a-Synuclein Mutants Created Y39F GFP Y125F GFP Y133F GFP S87A GFP S129A GFP Glycosylation sites unknown Two Stories Chapter 1: Characterizing The Newly Discovered E46K Mutant Chapter 2: Role of Post-Translational Modifications in aSynuclein E46K: Hypotheses and Aims Hypothesis 1. Expression of E46K a-synuclein will misfold, aggregate, and be toxic to yeast. Aims 1. Construct E46K mutant 2. Express wild-type and familial mutant E46K a-synuclein in S. cerevisiae yeast model. 3. Evaluate cellular localization and toxicity of wild-type versus E46K familial mutant form of a-synuclein expressed in S. cerevisiae. Site-Directed Mutagenesis Aim 1: Construction of E46K Mutant Methylated plasmid Methylation Mutagenesis X WT gene Primers: 1 contains target mutation X X X X Transformation into E. Coli Mutated plasmid -Glu residues were mutated to Lys (E K) Western Analysis Aim 2: Expression of E46K Mutant Heat to separate proteins Transfer Proteins Incubate Blot with Anti-bodies Development of Blot Visualization of Proteins Aim 2: Expression of E46K Western Analysis 148 E46K 98 64 ~124 kDa ~62 kDa 50 Predictions 36 GFP ~34kDa 22 16 MW Marker -E46K a-synuclein will have SDS insoluble aggregates -Dimer formation of E46K a-synuclein will be visualized Results: Expression of Familial Mutant E46K Western Blot + — +— +— + — + — + 98 148 64 ~62kDa 98 64 50 ~62kDa 36 50 ~34kDa ~34kDa 36 ~28kDa kDa MW 22 Sharma, 2004. Coomassie Stain -E46K runs 8-10 kDa higher than predicted -Lack of SDS insoluble aggregates Aim 3: Examining Toxicity of aSynuclein Predictions Optical Density and Spotting Growth Analyses Familial mutant a-synuclein will be toxic to yeast cells E46K mutant a-synuclein will be the most toxic to yeast cells Wild-type a-synuclein will not be toxic to yeast cells Results: E46K Mutant a-Synuclein Expression Is Toxic To Yeast Growth Curve 3 Log Cell Concentration 2.5 pYES2 2 GFP WT 1.5 A30P A53T A53T/ A30P 1 E46K 0.5 0 0 10 20 30 40 50 Time (hours) E46K expressing cells show a major lag in growth Results: E46K Mutant a-Synuclein Expression Is Toxic To Yeast Spotting 5X Less 5X Less 5X Less Glucose (non-inducing) Galactose (inducing) Parent Vector GFP WT E46K A30P A53T E46K expressing cells show no major decrease in growth rates Aim 3: Localization of E46K Predictions Live Cell GFP Microscopy E46K-GFP(CT) -E46K a-synuclein expression= foci formation -Localization to plasma membrane Results: a-Synuclein Localizes to the Periphery & Forms Foci Live Cell GFP Microscopy Wt-GFP E46K-GFP A30P-GFP A53T-GFP A30P/A53T-GFP - Halos are preserved -E46K shows increase foci formation compared to other familial mutants a-Synuclein Misfolding & Aggregation In vivo a-Synuclein Folding Live Cell Microscopy Wild-type a-Synuclein Increased Foci Formation No Toxicity Toxicity Toxicity Misfolded E46K a-Synuclein Increased Foci Formation Chapter 2 Role of Post-Translational Modifications in a-Synuclein Post-Translational: Hypotheses & Aims Hypothesis 1. Post-translational modifications of a-synuclein will decrease its misfolding and aggregation. 2. Expression of post-translational mutant a-synuclein will not be toxic to yeast. Aims 1. Construct post-translational S129A, Y39F, and Y125 mutants 2. Express wild-type and mutant S129A, Y39F, and Y125 a-synuclein in S. cerevisiae yeast model. 3. Evaluate cellular localization and toxicity of wild-type versus mutant forms of a-synuclein expressed in S. cerevisiae. Aim 2: Expression of a-Synuclein Predictions Western Analysis 148 98 WT ~62 kDa 64 Y125F 50 Y39F ~54 kDa S129A 36 GFP ~34kDa 22 16 MW Marker -Post-translational mutants will migrate at lower molecular weights -WT a-synuclein will run at ~62 kDa -Protein expression will be equal in all lanes Results: a-Synuclein Expression of S129A, Y39F, and Y125F Mutants Western Blot 148 98 64 ~62 kDa 50 ~34kDa 36 kDa MW Coomassie Stain -Surprisingly post-translational mutants run 8-10 kDa higher than predicted -Lack of SDS insoluble aggregates Aim 3: Examining Toxicity of aSynuclein Predictions Optical Density and Spotting: Growth Analysis S129A, Y39F, & Y125F mutant a-synuclein will not be toxic to yeast cells Wild-type a-synuclein will not be toxic to yeast cells Results: S129A, Y39F, and Y125F Mutant aSynuclein Expression Is Toxic To Yeast Growth Curve 3 Log Cell Concentration 2.5 2 S129A Y39F 1.5 WT Y125F 1 0.5 0 0 10 20 30 40 50 Time (hours) - Post-translational mutants show major growth deficiencies a-Synuclein Expression of S129A, Y39F, and Y125F mutants Spotting Non-inducing Inducing Parent Vector GFP WT Y39F Y125F S129A - Post-translational mutants show minor growth deficiencies Aim 3: Localization of a-Synuclein Mutants Predictions Live Cell GFP Microscopy S129A-GFP(CT) Y39F-GFP(CT) Y125F-GFP(CT) -Post-translational mutant a-synuclein will localize to plasma membrane Results: S129A, Y39F, and Y125F Mutant aSynuclein Localizes Near Yeast Plasma Membranes Live Cell GFP Microscopy GFP Y39F-GFP Y125F-GFP S129A-GFP Wt-GFP - Halos are preserved -Post-translational modifications show lack of foci formation Conclusions 1. Familial E46K mutant a-synuclein induces toxicity upon expression 2. Increased foci formation with E46K a-synuclein expression 3. a-Synuclein’s increased size in not due to phosphorylation at Serine 129 and nitrosylation at Tyrosines 39 and 125 4. S129A, Y39F, and Y125F mutant a-synuclein showed unexpected increase in toxicity 5. In vivo membrane association of S129A, Y39F, and Y125F a-synuclein Discussion E46K Toxicity May Be Related To Increased Misfolding Zarranz, et al., 2004: Study showed that E46K a-syn is more prone to aggregation compared to other familial mutants E46K had extensive peripheral localization and increased foci formation compared to other a-syn expressing cells OD600 showed that E46K cells have large lag in growth; spotting assays show no inhibited growth rate. Increased aggregation of E46K a-syn may increase its toxicity = cell death Discussion Increased Size: Not Due to Phosphorylation or Nitrosylation DebBurman yeast model: a-syn ran ~8-10 kDa higher a-Syn migrated higher than predicted due to post-translation modifications on Ser129 & Tyr 39 and 125 No change in migration patterns of a-syn deficient for these residues Increased size not due to phosphorylation or nitrosylation Increased size maybe due to other modifications Discussion Post-translational Mutants Showed Unexpected Increase In Toxicity Giasson, et al., 2002: nitrosylation and phosphorylation modifications may be responsible for inclusions seen in PD patients Formation of inclusions coincides with disease onset We expected to see less toxicity when key sites are mutated Phosphorylation or nitrosylation modifications maybe beneficial to a-syn expressing cells Discussion In vivo membrane association of S129A, Y39F, and Y125F a-Synuclein DebBurman yeast model: Peripheral localization of wild-type a-syn Post-Translational mutant a-syn localized to yeast plasma membrane a-Syn contains a motif that has the ability to bind phospholipids vesicles The cytoplasm of yeast cells is smaller than those in neurons; a-syn may have easier ability to bind to membranes Future Studies 1. Examine other a-synuclein residues linked to nitrosylation and phosphorylation sites. 2. Examine other post-translational modification sites linked to a-synuclein misfolding. 3. Assessment of stability of mutant forms of a-synuclein in S. cerevisiae. Acknowledgements DebBurman Lab Dr. Shubhik DebBurman Isaac Holmes Nijee Sharma Katrina Brandis Ruja Shrestha Lavinia Sintean Tasneem Saylawala Arun George Paul Jessica Price NIH NSF