* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download U(x) - Bioconductor
Survey
Document related concepts
Transcript
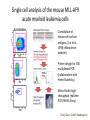
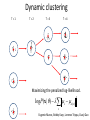
Analysis of Gene Expression at the Single-Cell Level Guo-Cheng Yuan Department of Biostatistics and Computational Biology Dana-Farber Cancer Institute Harvard School of Public Health Bioconductor, July 31st, 2014 bioconductor Methods to sequence the DNA and RNA of single cells are poised to transform many areas of biology and medicine. --- Nature Methods • “Recent technical advances have enabled RNA sequencing (RNA-seq) in single cells. Exploratory studies have already led to insights into the dynamics of differentiation, cellular responses to stimulation and the stochastic nature of transcription. We are entering an era of single-cell transcriptomics that holds promise to substantially impact biology and medicine.” – R. Sandberg, 2014. Nature Methods Cell Division Cell-type A Cell-type C Cell-type B Cell-type E Cell-type D Cell-type F R. Sandberg, 2014. Nature Methods Challenges in single-cell data analysis • Characterize and distinguish technical/biological variability • Identify new and meaningful cell clusters. • Identify the lineage relationship between different cell clusters. • Characterize the dynamic process during cellstate transitions. • Elucidate the transition of regulatory networks. • Distinguish stochastic vs real variation CMP GMP MEP MEP CLP Guoji Guo, Eugenio Marco SPADE: a density-normalized, spanning tree model Down-sample Clustering, Spanning-tree Visualization Qiu et al. 2011 Nat Biotech, p886 Log2(CMP1/CMP2) CD55 7.87 ICAM4 3.98 CD274 3.32 MPL 3.19 TEK 2.83 Cancer Stem Cells • Each cancer contains a highly heterogeneous cell population. • Clonal evolution contributes to cancer heterogeneity • Cancer cells are hierarchically organized and maintained by cancer stem cells • How are the leukemia stem cells related to normal blood cell lineage? How do they differ? Single cell analysis of the mouse MLL-AF9 acute myeloid leukemia cells Compilation of mouse cell surface antigens (Lai et al., 1998; eBioscience website) Primer design for 300 multiplexed PCR (collaboration with Helen Skaletsky) Micro-fluidic highthroughput realtime PCR (96.96 Array) Guoji Guo, Assieh Saadatpour t-SNE analysis identifies similarities between cell-types • t-SNE is a nonlinear dimension reduction method, and can identify patterns undetectable by PCA • t-SNE minimizes the divergence between distributions over pairs of points. • Leukemia cells are more similar to GMPs than to HSCs • Leukemia cells are highly heterogeneous. Mapping leukemia cells to normal hematopoietic cell hierarchy • Use 33 common genes to map cell hierarchy. • Mapping identifies two subtypes of leukemia cells. • These cells are similar but not identical to their corresponding normal lineages. Coexpression networks are different among subtypes All Leukemia Leukemia 1 GMP Leukemia 2 Surani and Tischler, Nature 2012 Guo et al. Dev Cell 2010 Dynamic clustering T=1 T=2 T=3 T=4 Maximizing the penalized log-likelihood. log P(x | ) c a(c) 2 c Eugenio Marco, Bobby Karp, Lorenzo Trippa, Guoji Guo Identifying bifurcation points and directions EPI ICM PE TE >80% variance increase during bifurcation is attributed to a single (bifurcation) direction. Modeling dynamics by bifurcation analysis dx U ( x) dt I) U(x) 4a 3 27b 2 0 II) U(x) 4a 3 27b 2 0 x 4 ax 2 U ( x) bx 4 2 Modeling dynamics by bifurcation analysis dx U ( x) dt dW (t ) I) U(x) 4a 3 27b 2 0 II) U(x) 4a 3 27b 2 0 Noise level has large impact on lineage biases =1 = 0.5 =2 Lineage bias due to perturbation of TF activity Control U(x) Perturbation U(x) Predicted lineage bias due to 2 fold decrease of TF level Experimental validation using Nanog mutant PE EPI Nanog How do we infer dynamics without temporal information? Characterization of early bipotential progeny of Lgr5+ intestinal stem cells Crosnier 2006. Nature Review Tae-Hee Kim, Assieh Saadatpour Principal Curve Analysis Reconstruct Temporal Information t-SNE plot indicates two distinct clusters, linked a small number of transitional cells Principal Curve Analysis Reconstruct Temporal Information t-SNE plot indicates two distinct clusters, linked a small number of transitional cells Principal curve analysis captures the overall trend of cell-state transition Olfm4 Rnf43 apdh dca7 Actb Pcna Taf1d oc2-2 Cd24 Itgav Vil1 rom1 CD44 Gcnt3 t1h1e Axin2 oc2-1 ox9-1 Cnn3 Cdx2 Ccl9 csm3 Kitl Gpld1 Vdr Hopx es1-2 Fgfr4 dkn1b Nr4a2 Vegfc Tex9 Znrf3 Lrp1 Cd9 Dpp4 Farp1 Ccnl1 Ascl2 ox9-2 c14a1 Lrig1 Nop2 cnd1 Lcp1 Hprt Ly75 Il6st obtb3 Cdx1 Tyms Bmi1 Fos Nfat5 Atf3 nase1 Dll1 Cd14 Zfp36 Cd82 emin4 Sycn Hig2 Il6 Rfc4 Cdk1 Defa5 cnb1 Rrm2 Plk1 Ang4 asp12 Tlr1 Lct Kit Apoa1 Lifr Tdgf1 Dct Muc2 Spdef Nupr1 Dll4 Tert-1 urod1 Tert-2 Clca3 Lipe toh1-1 Tbx3 usp1 JunB Psrc1 dkn2b Bmp4 Hes3 Treh ep15 Chga Alpi Pax4 toh1-2 Cck dkn1a urog3 cna2 Mki67 Defa3 achd1 Ephb3 Fstl1 enpe hisa2 Nfatc3 Jun Prelp otch1 es1-1 Fgfr2 es1d Apex1 Abcg2 Igf1r Lgr5 Agr3 Arg2 ap1l1 Clca1 Heph ma4d otch2 dr90 Ppif Tac1 Egr2 cne1 Selm Gas6 Sox4 Hbegf Cd38 Fosb Dbd2 Lyz1 Clca2 Igfbp4 asa3 Msi Cd55 Aspa Ifnar1 Tcof1 usp4 Mmp7 Kcnq1 Syp Dclk1 Chgb d51l1 GFP Hes5 E2F1 Egr1 Hey1 Insm1 m614 Hey2 Sp5 Btla Lyz2 Heyl Ccl6 Reg4 Myc Hes7 reb3l4 Cd83 Fcgbp dkn1c Zic2 cam2 Gfi1 kx2.2 md1a Gfi1b dkn2a Tff3 Diap3 Cdkn3 Inferred dynamic gene expression profile Use the principal curve coordinate as a proxy for temporal evolution. 10 9 8 7 6 5 4 3 2 1 Conclusions • Single-cell genomics is a powerful technology for understanding cellular heterogeneity and hierarchy. • Single-cell gene expression data analysis present many new methodological challenges. • It is a great time to develop algorithms and software for single cell data analysis. Acknowledgement Eugenio Marco Assieh Saadatpour Stuart Orkin Guoji Guo Bobby Karp Ramesh Shivdasani Tae-Hee Kim Lorenzo Trippa Paul Robson Funding from NIH, HSCI