* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Enzymology Part 2
Restriction enzyme wikipedia , lookup
Inositol-trisphosphate 3-kinase wikipedia , lookup
Alcohol dehydrogenase wikipedia , lookup
Multi-state modeling of biomolecules wikipedia , lookup
Lactoylglutathione lyase wikipedia , lookup
Beta-lactamase wikipedia , lookup
Cooperative binding wikipedia , lookup
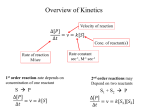
Enzymology Part 2 PRINCIPLES OF ENZYMOLOGY TRANSITION STATE THEORY: Colliding molecules of the reactants must have sufficient energy to overcome a potential energy barrier (the activation energy) to react Factors affecting rate of a rxn: a. Sustrate concentration b. Temperature c. pH d. Inihibitors, effectors A. SUBSTRATE CONCENTRATION A B PART A ([S] is low while [E] is high) • • • Directly proportional Initial velocity (0) increases as [S] is small compared to [E]) If [E] is higher than [S] in this section of the graph, the frequency of collision is high PART B ([S] is high while [E] is low • • • • Rate no longer depends on [S] [S] higher than [E] Active site is saturated with substrate Frequency of collision no longer the determining factor Determination of 0 All the test tubes have the same contents except that each has a different substrate concentration Either monitor the disappearance of substrate or the formation of product. Therefore can obtain the rate of disappearance of S or formation of P at each substrate concentration 0 Vmax = maximum velocity (achieved when all the substrate molecules complex with E or enzymes are saturated with substrates) • Km & Vmax are constants which are unique for a pair of enzyme and its substrate Km = initial velocity = Michaelis constant (takes into account all the rxn constants of the rxns involved k1 E+S k3 ES k2 • E+P Km = (k2+k3)/k1 k4 Km = substrate concentration at ½Vmax • Units of Km = dm-3 • Vmax [S] o = -------------Km + [S] MICHAELIS MENTEN EQUATION • Km can be determined graphically • Km shows the affinity of an enzyme for a substrate • Km & Ks inversely related • The bigger the value of Km, the lower the affinity • Small Km value, the higher the affinity LINEWEAVER-BURK PLOT Km and Vmax can be determined from the vs [S] The Michaelis Menten equation can be modified to obtain a more Km and Vmax precise values 1/0 = [Km/Vmax]. 1/[S] + 1/Vmax ENZYME ACTION CAN BE CONTROLLED BY: Non covalent inhibition: 2. Competitive inhibition 3. Non competitive inhibition 4. Uncompetitive inhibit 5. Covalent inhibition: Irreversible 6. Allosteric Control COMPETITIVE INHIBITION Inhibitor competes with the substrate for the active site of the same enzyme Inhibitor and substrate have similar chemical structures Lack of specificity at the active site: Cannot differentiate inhibitor from substrate Inhibitory effect can be overcome by increasing substrate concentration KINETICS OF COMPETITIVE INHIBITION Note that: Vmax (- I) denoted V’max (in the presence of Competitve I) Km < K’m Because I and S compete for the same site. More S is required to achieve half saturation point. Hence Km increases. K’m is more than Km by a factor of (I + [I]/Ki) V’maks = Vmaks (I + [I]/Ki) When enzyme is saturated with I, the effect can be overcome by increasing [S]. So when the enzyme becomes saturated with the S then Vmax is achieved. Example: inhibition of folic acid synthesis by sulphanamide PABA (para amino benzoic acid Sulphanamide PABA is required for the synthesis of folic acid Sulphanamida is a drug that competes with PABA for the enzymes in the folic acid synthesis pathway. Find another example of competitive inhibition in the cell Non-Competitive Inhibition 1. The inhibitory effects cannot be overcome by increase in Substrate concentration 2. Inhibitor binds to a site other than the active site. Binding of I does not affect binding of S 3. Therefore in this case, structure of inhibitor is not similar to substrate 4. Inhibitory effects depend on I and Ki and not on [S] 5. I can bind to either E or ES I EI Gradient = Km/Vmax I/V’max = (I + [I]/KI)(I/Vmax Vmax reduces in the presence of Km does not change non competitive inhibitor. It is as though the amount of E is now less Uncompetitive Inhibition 1. Binds only to the ES complex but not the free enzyme 2. Increasing [S] will increase the [ES] thus increasing [S] will not reverse the effects of an uncompetitive inhibitor. 3. Lineweaver Burke plots will give a set of parallel lines Note: Both intercepts change but slope remains constant Irreversible Inhibition 1. Enzymes can be inhibited by an irreversible manner for example by covalent attachment either to E or ES 2. Kinetic pattern looks like non-competitive inhibition (net effect is a loss of active enzyme): Vmax decreases 3. Reaction is time dependent decrease in enzymatic activity ie not instantaneous as seen in non competitive inhibition 4. Penicillin is an irreversible inhibitor: binds to serine residue in the active site of a the enzyme (glycoprotein peptidase). Affects cell wall synthesis, making bacterial cells susceptible to rupture. 5. Others include: Hg2+, Pb2+, arsenic 6. Binds to functional groups such as: –COOH, -NH2, -SH dan -OH Effect of Temperature: Temperature increases the rate of reaction Reaction rates increase because of the increase in collision/min between the substrates BUT at extreme temperatures, enzyme activity decreases because of enzyme denaturation Effect of pH • Influences ionisation of functional groups in proteins • As pH changes, the ionisation state of the functional groups of the amino acids in the tertiary structure and in the active site will change affecting enzyme activity ENZYME REGULATION Enzyme activity can be regulated by: 1. Covalent modificatio: eg phosphorylation of protein kinase 2. Zymogens 3. Allosteric regulation: non covalent interaction between enzymes and small molecules (metabolites) In allosteric regulation enzyme activities controlled at key steps in metabolic pathways: Feedback inhibition (feedback regulation) The enzyme involved is a regulatory enzyme Characteristics of Allosteric Enzymes 1. They have 2 binding sites: the active site and the modulator site 2. High molecular weight 3. Very complex 4. Difficult to purify 5. Contains 2 or more polypeptide chains (subunits): more than 1 S- binding site/enzyme molecule 6. Does not obey Michaelis Menten kinetics: vs [S] yields a sigmoid graph compared to a hyperbolic curve The binding of 1 substrate to a protein molecule makes it easier for additional substrate molecules to bind to the same protein: substrate binding is termed cooperative Characteristics of Allosteric Enzymes (con’t…) 6. Allosteric enzymes are regulated by activation: ie there are effectors or modulators that can have a positive (stimulatory) or negative effects on enzyme activity POSITIVE COOPERATIVE EFFECT and NEGATIVE COOPERATIVE EFFECT These enzymes take some time to reach saturation point. MODEL TO EXPLAIN ALLOSTERIC ENZYME KINETICS 1. SYMMETRY MODEL: Monod, Wyman and Changeux a. Allosteric proteins exist in two conformational stages: R = Relaxed (high affinity for substrate) and T = Taut (low affinity for substrate b. Model named symmetry because in each protein molecule all the subunits have either R or T conformation c. These 2 states are in equilibrium R0 T0 1. SYMMETRY MODEL: Monod, Wyman aand Changeux (con’t…) d. Presence of S will result in binding to R0 to form R1. This reduces the R0 concentration of disturbing theT0/R0 equilibrium. To restore equilibrium, molecules in the T0 conformation will change to the conformation R0 e. Positive homotropic effectors f. This model also provides for binding to positive and negative effectors. If effctors are not substrates then they are known as heterotropic effectors g. Effectors that promote Substrate binding are known as positive heterotropic effectors h. Effectors that diminish Substrate binding are known as negative heterotropic effectors i. Positive effectors increase number of available binding sites j. Negative effectors decrease number of available binding sites 2. SEQUENTIAL MODEL: Koshland, Nemethy and Filmer (Involves cooperativity and Conformational Changes) Basis: 1. Protein molecules are not in symmetry (ie asymmetric). 2. Proteins are flexible molecules and conformations altered when ligands bind 3. Binding of a ligand to one subunit of a multimeric protein, would cause conformational changes to occur, and then through contacts with the other subunits cause their conformation to change. 4. As a consequence other subunits will have either greater or lesser affinity for the ligand Aspartate Transcarbomylase