* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Lecture 31
Survey
Document related concepts
Transcript
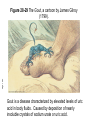
FCH 532 Lecture 31 Chapter 28: Nucleotide metabolism Quiz on Mon (4/16): IMP synthesis-Purine synthesis Quiz on Wed(4/18): Pyrimidine biosynthesis/regulation Quiz on Friday(4/20): Ribonucleotide reductase mechanism Friday (4/20): extra credit seminar, Dr. Jimmy Hougland, 145 Baker, 3-4PM. ACS exam has been moved to Monday (4/30) Quiz on Final is scheduled for May 4, 12:45PM-2:45PM, in 111 Marshall Page 1077 Page 1080 Proposed mechanism for rNDP reductase Proposed reaction mechanism for ribonucleotide reductase 1. Free radical abstracts H from C3’ 2. Acid-catalyzed cleavage of the C2’-OH bond 3. Radical mediates stabilizationof the C2’ cation (unshared electron pair) 4. Radical-cation intermediate is reduced by redoxactive sulhydryl pairdeoxynucleotide radical 5. 3’ radical reabstracts the H atom from the protein to restore the enzyme to the radical state. Catabolism of purines • • • All pathways lead to formation of uric acid. Intermediates could be intercepted into salvage pathways. 1st reaction is the nucleotidase and second is catalyzed by purine nucleoside phosphorylase (PNP) • Ribose-1-phosphate is isomerized by phosphoribomutase to ribose-5-phosphate (precursor to PRPP). Purine nucleoside + Pi • Purine base + ribose-1-P Purine Adenosine and deoxyadenosine are notnucleoside degraded by PNP but are deaminated by phosphorylase adenosine deaminase (ADA) and AMP deaminase in mammals Figure 28-23 Major pathways of purine catabolism in animals. ADA Page 1093 Genetic defects in ADA kill lymphocytes and result in severe combined immunodeficiencey disese (SCID). No ADA results in high levels of dATP that inhibit ribonucleotide reductase-no other dNTPs Page 1094 Figure 28-24a Structure and mechanism of adenosine deaminase. (a) A ribbon diagram of murine adenosine deaminase in complex with its transition state analog HDPR. Page 1094 Figure 28-24b (b) The proposed catalytic mechanism of adenosine deaminase. 1. Zn2+ polarized H2O molecule nucleophilically attacks C6 of the adenosine. His is general base catalyst, Glu is general acid, and Asp orients water. 2. Results in tetrahedral intermediate which decomposes by elimination of ammonia. 3. Product is inosine in enol form (assumes dominant keto form upon release from enzyme). Purine nucleotide cycle • • • Deamination of AMP to IMP combined with synthesis of AMP from IMP results in deaminating Asp to yield fumarate. Important role in skeletal muscle-increased activity requires increased activity in the citric acid cycle. Muscle replenishes citric acid cycle intermediates through the purine nucleotide cycle. Page 1095 Figure 28-25 The purine nucleotide cycle. Xanthine oxidase • • • • • • Xanthine oxidse (XO) converts hypoxanthine to xanthine, and xanthine to uric acid. In mammals, found in the liver and small intestine mucosa XO is a homodimer with FAD, two [2Fe-2S] clusters and a molybdopterin complex (Mo-pt) that cycles between Mol (VI) and Mol (IV) oxidation states. Final electron acceptor is O2 which is converted to H2O2 XO is cleaved into 3 segments. The uncleaved enzyme is known as xanthine dehydrogenase (uses NAD+ as an electron acceptor where XO does not). XO hydroxylates hypoxanthine at its C2 position and xanthine at the C8 positon to produce uric acid in the enol form. Figure 28-26a X-Ray structure of xanthine oxidase from cow’s milk in complex with salicylic acid. N-terminal domain is cyan Page 1095 Central domain is gold C-terminal domain is lavender Mechanism for XO 1. Reaction initiated by attack of enzyme nucleophile on the C8 position of xanthine. 2. The C8-H atom is eliminated as a hydride ion that combines with Mo (VI) complex, reducing it to Mo (IV). 3. Water displaces the enzyme nucleophile producing uric acid. Page 1096 Figure 28-27 Mechanism of xanthine oxidase. Figure 28-23 Major pathways of purine catabolism in animals. ADA Page 1093 Genetic defects in ADA kill lymphocytes and result in severe combined immunodeficiencey disese (SCID). No ADA results in high levels of dATP that inhibit ribonucleotide reductase-no other dNTPs Purine degredation in other animals Primates, birds, reptiles, insects-final degradation product id uric acid which is excreted in urine. Goal is the conservation of water. Page 1097 Figure 28-29 The Gout, a cartoon by James Gilroy (1799). Gout is a disease characterized by elevated levels of uric acid in body fluids. Caused by deposition of nearly insoluble crystals of sodium urate or uric acid. Clinical disorders of purine metabolism Excessive accumulation of uric acid: Gout The three defects shown each result in elevated de novo purine biosynthesis Common treatment for gout: allopurinol Allopurinol is an analogue of hypoxanthine that strongly inhibits xanthine oxidase. Xanthine and hypoxanthine, which are soluble, are accumulated and excreted. Catabolism of pyrimidines • • • Animal cells degrade pyrimidines to their component bases. Happen through dephosphorylation, deamination, and glycosidic bond cleavage. Uracil and thymine broken down by reduction (vs. oxidation in purine catabolism). Page 1098 Biosynthesis of of NAD and NADP+ Page 1099 Produced from vitamin precursors Nicotinate and Nicotinamide and from quinolinate, a Trp degradation product Biosynthesis of FMN and FAD from riboflavin Page 1100 FAD is synthesized from riboflavin in a tworeaction pathway. Flavokinase phosphorylates the 5’OH group to give FMN FAD pyrophosphorylase catalyzes the next step (coupling of FMN to ADP). Page 1101 Biosynthesis of CoA from pantothenate