* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Document
Survey
Document related concepts
Transcript
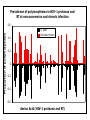
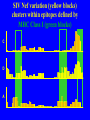
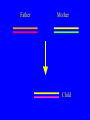
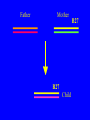
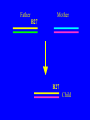
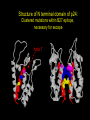
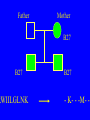
“What limit can be put to this power (natural selection), acting during long ages and rigidly scrutinising the whole constitution, structure, and habits of each creature,-favouring the good and rejecting the bad.” Charles Darwin Origin of the Species Natural Selection • All species produce more offspring than can possibly survive and reproduce. • Organisms differ in their ability to survive and reproduce, in part due to differences in genotype. • In every generation, those genotypes that promote survival in the current environment are present in excess at the reproductive age and therefore contribute disproportionately to the offspring of the next generation. • For natural selection to operate there must be heritable variation for traits that are correlated with reproductive success. Average Rates of Nucleotide Substitution in Different Organisms Organism/Genome Plant chloroplast DNA Mammalian nuclear DNA Plant nuclear DNA E. coli and Salmonella enterica bacteria Drosophila nuclear DNA Mammalian mitochondrial DNA HIV-1 Substitution Rate (per site, per year) ~ 1 x 10-9 3.5 x 10-9 ~ 5 x 10-9 ~5 x 10-9 1.5 x 10-8 5.7 x 10-8 6.6 x 10-3 The Degeneracy of the Genetic Code Proline (4 synonymous codons) CCT CCC CCA CCG Histidine (2 synonymous codons) CAT CAC CAA - Gln CAG - Gln Measuring Selection Pressures in Genes • Selection pressures can be measured by comparing the relative rates (ratio) of synonymous (silent) (dS) and nonsynonymous (amino acid changing) (dN) substitutions: Seq 1: Seq 2: Ser TCA † TCG Ser Met ATG * ATA Ile Leu TTA † CTA Leu Gly GGG † GGT Gly Gly GGA ** ATA Ile † Synonymous substitution * Nonsynonymous substitution dN/dS < 1.0 = negative selection (functional constraint - most genes) dN/dS ~ 1.0 = neutral evolution (pseudogenes) dN/dS > 1.0 = positive selection Genetic Variability of HIV • HIV exhibits extremely high levels of genetic variation: the virus evolves about 1 million times faster than human DNA (HIV-1 = ~10-3 subs/site/year, human DNA = 10-9 subs/site/year) . • This is mainly due to: – Extremely high mutation rate, particularly because the replication enzyme reverse transcriptase that converts RNA to DNA is very error prone – High turnover of virus within infected individual ~1010 viruses produced in each patient each day – Rapid generation time (~2.6 days) Phylogeny of global HIV-1 isolates + 197 strains from the Congo Genetic diversity at transmission • Early analyses concentrated on the env gene and revealed a low level of diversity in seroconverters • Ho D., Science, Leigh-Brown A., J. Virol • So the infection was thought to be established by a small “founder” set of clones • But that work did produce evidence of greater diversity in gag, nef and pol than env Four Sexual Transmitters Subtype A D1 1.31 R1 D2 Subtype B Gag p24 Neighbour-Joining Tree 1.28 R2 D3 1.05 R3 D4 Donor Recipient Subtype Reference sequences 1.0 Percent Divergence 1.02 R4 P32-Ia-M24 P32-Ia-M08 P32-Ia-J05 P32-Ia-M19 P32-Ia-M14 P32-Ia-M09 P32-Ia-J16 P32-Ia-J17 P32-Ia-M04 P32-Ia-J03 P32-Ia-M23 P32-Ia-M18 P32-Ia-M16 P32-Ia-M11 P32-Ia-M07 P32-Ia-M05 P32-Ia-M21 P32-Ia-M22 P32-Ia-M06 P32-Ia-M20 P32-Ia-M13 P32-Ia-J04 P32-Ia-J06 P32-Ia-M12 P32-Ia-M10 P32-Ia-M25 P32-Ia-J02 P32-Ia-J11 P32-Ia-J10 P32-Ia-J08 P32-Ia-J07 P32-Ia-J09 P32-Ia-J12 P32-Ia-J14 P32-Ia-M17 P32-Ia-M01 P32-Ia-M02 S32-I-A04 S32-Ia-AP11 S32-Ia-AP10 S32-Ia-AP06 S32-Ia-AP04 S32-I-AP12 S32-I-AP11 S32-I-AP07 S32-I-AP03 S32-I-AP01 S32-Ia-O10 S32-Ia-O05 S32-Ia-O02 S32-I-O11 S32-I-O10 S32-I-O08 S32-I-O07 S32-I-O06 S32-I-O05 S32-I-O04 S32-I-O03 S32-I-O01 S32-I-A18 S32-I-A17 S32-I-A15 S32-I-A14 S32-I-A13 S32-I-A12 S32-I-A11 S32-I-A08 S32-I-A06 S32-I-A05 S32-I-AP02 S32-I-AP06 S32-Ia-O07 S32-Ia-O12 S32-I-O02 S32-I-O09 S32-Ia-O01 S32-Ia-O04 S32-Ia-O03 S32-I-AP04 S32-Ia-AP12 S32-I-AP10 S32-I-AP08 S32-I-AP05 S32-I-AP09 S32-Ia-AP09 S32-Ia-AP08 S32-Ia-AP07 S32-Ia-AP03 S32-Ia-AP02 S32-Ia-AP05 0.01 p17-a-018.p47 p17-a-020.p47 p17-a-010.p47 p17-a-019.p47 p17-a-012.p47 p17-a-011.p47 p17-a-022.p47 p17-a-015.p47 p17-a-008.p47 p17-a-048.p47 p17-a-047.p47 p17-a-046.p47 p17-a-045.p47 p17-a-044.p47 p17-a-043.p47 p17-a-042.p47 p17-a-040.p47 p17-a-039.p47 p17-a-037.p47 p17-a-036.p47 p17-a-035.p47 p17-a-033.p47 p17-a-032.p47 p17-a-028.p47 p17-a-025.p47 p17-a-026.p47 p17-a-041.p47 p17-a-038.p47 p17-a-034.p47 p17-a-031.p47 p17-a-030.p47 p17-a-029.p47 p17-a-027.p47 p17-a-001.p47 p17-a-024.p47 p17-a-023.p47 p17-a-021.p47 p17-a-017.p47 p17-a-016.p47 p17-a-013.p47 p17-a-009.p47 p17-a-007.p47 p17-a-006.p47 p17-a-005.p47 p17-a-004.p47 p17-a-002.p47 p17-a-003.p47 p17-a-014.p47 DONOR RECIPIENT UNRELATED SEROCONVERTER Maximum Likelihood phylogenetic tree Prevalence of polymorphisms in HIV-1 protease and RT at seroconversion and chronic infection Proportion of polymorphisms 0.6 >1 year Seroconversion 0.4 0.2 0 1 26 51 76 RT2 RT2 7 RT5 2 RT7 7 RT1 02 RT1 2 7 RT1 5 2 -0.2 -0.4 -0.6 Amino Acid (HIV-1 protease and RT) RT1 7 7 RT2 02 RT2 2 7 Evading Cytotoxic T-Lymphocyte Recognition May Enhance Viral Fitness Evading Cytotoxic T-Lymphocyte Recognition May Enhance Viral Fitness Selection of escape mutants A replication A1-A101 SELECTION PRESSURE e.g. A6 Some lead to increased fitness - survive e.g. A12 Some mutations lead to reduced fitness - die off Continued selection of escape mutants A6 replication SELECTION PRESSURE A6.1-A6.101 e.g.A6.4 e.g. A6.9 Some lead to increased Some mutations lead to reduced fitness - survival fitness - extinction “Extinction has only separated groups: it has by no means made them: for if every form which has ever been were suddenly to reappear…all would blend together by steps as fine as those between the finest existing varieties” Charles Darwin Origin of the Species Viral Load CTL Response Do CTL Select for Escape Variants During Acute Infection? Weeks Years Rev Tat Vpx Tat Gag Vpr Pol 700 1400 2100 Rev Vif 2800 3500 4200 4900 5600 6300 Nef Env 7000 7700 8400 9100 9800 10500 5 CTL Epitopes Accumulate Variation at Different Rates During Acute SIV Infection Consensus QGQYMNTPW ARRHRILDIYL IRFPKTFGW GDYKLVEI KRQQELLRL Inoculum ......... ........... ......... ........ ......... ......... ........... T.Y...... ........ ......... ...H..... ........... T.Y...... ........ ......... ......... E.......... T.Y...... ........ ......... ......N.. E.......T.. T.Y..I... ........ ......... ......N.. E.......M.F T.Y..I... ........ ......... ......N.. E.......K.. T.Y..I... ........ ......... ....I.N.. E.......K.. ..Y..I... .....I.V ...H..... ......N.. E.......K.. T.Y..I... .....I.V ...H..... Epitope Variants Reduce CTL Recognition G are Qraphics uickTim needed decom e™ toand see pressor athis picture. 100 Peptide bindingtoclassI CTLrecognition 60 40 20 IRFPKTFGW GDYKLVEI QGQYMNTPW KRQQELLRL ARRHRILDIYL 0 Wild-type %WildType 80 SIV Nef variation (yellow blocks) clusters within epitopes defined by MHC Class I (green blocks) G are Qraphics uickTim needed decom e™ toand see pressor athis picture. C G are Qraphics uickTim needed decom e™ toand see pressor athis picture. D G are Qraphics uickTim needed decom e™ toand see pressor athis picture. A Summary • Antiretrovirus T lymphocytes recognise peptide antigens dictated by, and bound to MHC Class I molecules • Genetic variants of the virus can alter or even abolish immune recognition of infected target cells • Viruses bearing these sequences have a survival advantage • Escape mutants grow out (at different rates) and are positively selected 26 year old man admitted with aseptic meningitis • • • • • • Had unprotected anal sex on 20 Sept.1995 HIV Elisa negative on 10 October 1995 P24 gag antigen positive Viral load >10 millions copies per ml CTL assay 19 October 1995 Single nef response HLA B8 NEF EPITOPE FLKEKGGL Intra-Host Evolution of HIV-1 SIHIGPGRAFYTTGE SIPIGPGRAFYTTGQ SIHIGPGGAFYTTGQ SIHIGPGRAFYTTGD SIPIGPGRAFYTTGD GIHIGPGSAFYATGD SIHIGPGRAFYTTGG SIHIGPGRAVYTTGQ GIHIGPGSAFYATGG GIHIGPGRAVYTTEQ RIHIGPGRAVYTTEQ GIHIGPGSAFYATGR RIYIGPGRAVYTTEQ GIHIGPGSAVYATGG RIYIGPGSAVYTTEQ GIHIGPGSAFYATGG RIGIGPGRSVYTAEQ GIHIGPGSAVYATGD GIHIGPGRAFYATGD GIHIGPGRAVYTTGD RIYIGPGRAVYTTDQ Tip of the V3 loop (part of the envelope protein of HIV-1) - diversity in a single patient • The HIV-1 envelope protein is under very strong positive selection to help the virus escape from the human immune response (the V3 loop contains epitopes for neutralising antibodies and cytotoxic T-lymphocytes (CTLs). • V3 loop dN/dS = 13.182 (Nielsen & Yang. Genetics 148, 929. 1998). Does transmission of CTL escape viruses impact on the global epidemic? Questions • Child can inherit a HLA Class I molecule which has dictated a response in the mother • The maternal immune response can select escape mutants Are these immune escape viruses detectable in the mother? Can the mother transmit these viruses? Do these escape viruses propagate infection in the child? Father Mother Child Father Mother B27 Child B27 Father B27 Mother B27 Child KK10 lies within a conserved region of p24 Gag 100% 50% 40% 20% 10% 5% 2% 1.3% P I V Q N L Q G Q MV H Q A I S P R T L N A WV K V V E E K A F S P E V I P M F S A L S E G A T P Q D L N T M L N T V G G H Q A A M Q M L K E T I N EEAAEWDRLHPVHAGPIAPGQMREPRGSDIAGTTSTLQEQIGWMTNNPPIPVGEIYKRWIILGLNKIVRMYSPTSILDIKQGPKEPFRDYVDRFYKTLRAEQASQEVKNWMTETLLVQNANPDCKTILKALGPAATLEEMMTACQGVGGPGHKARVL 1 4567 11 14 25 2729 41 44 48 53 71 79 83 86 90 92 96 98 107 110 114 120 123 128 148 154 159 KRWIILGLNK 167 170 177 183 187 194 199 208 213 216 219 223 230 Structure of N terminal domain of p24: Clustered mutations within B27 epitope, necessary for escape helix 7 Amino acid changes in the antigenic peptide at the second position prevent binding to HLA B27 molecules 100 % Maximum 80 KRWIILGLNK variants 60 KRWIILGLNK=R2L6 M =R2M6 K M =K2M6 K =K2L6 T M =T2M6 40 20 0 -9 -8 -7 -6 -5 peptide concentration (log molar) -4 The HLA B27 Gag peptide is not recognized in children of HLA B27-positive mothers WHY? 2000 1925 1600 IFN-g SFC/ 1200 million 800 PBMC 400 0 0 0 0 043-C 002-C 048-C 049-C B27-ve mother B27+ve mothers The HLA B27 bearing MOTHERS acquired an escape virus and transmitted it to the CHILDREN No P2 anchor mutation 2000 1600 IFN-g SFC/ 1200 million 800 PBMC 400 0 P2 anchor mutation shared with mother 0 0 0 043-C 002-C 048-C 049-C B27-ve mother B27+ve mothers Father Mother B27 B27 RWIILGLNK B27 - K- - -M- - HIV IN PERTH, AUSTRALIA Hypotheses If CTL-driven selection operates on HIV-1 in populations then selected mutations should: • be within or close to CTL epitopes • because of genetic restriction of immune responses, mutations should be linked to particular HLA variants • viral escape mutations would be evident as HLAassociated polymorphisms. HLA alleles significantly associated with HIV-RT polymorphisms within published epitopes putative epitopes flanking residues =Significant after correction Moore et al (2002) Science 296, 1439 Natural selection by AIDS Who wins the race? • HIV-1 10 subs/site/year -3 • Human genome (eg HLA) -9 10 subs/site/year “Nothing in biology makes sense, except in the light of evolution” Theodosius Dobzhansky 1973 Acknowledgments • • • • • • • • • • Eddie Holmes Paul Klenerman David Watkins Angela McLean Simon Mallal David O’Connor David Price Annette Oxenius Andrew McMichael Charles Bangham • • • • • • • • Philippa Easterbrook Jonathan Weber Sarah Fidler George Scullard Anele Waters Anne Edwards Philip Goulder The patients of The Chelsea and Westminster Hospital; St Mary’s Hospital; KwaZulu Natal