* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Pharmacogenetic Testing Prior to Initiation of Warfarin
Survey
Document related concepts
Neuropharmacology wikipedia , lookup
Discovery and development of direct Xa inhibitors wikipedia , lookup
Pharmaceutical industry wikipedia , lookup
Prescription costs wikipedia , lookup
Pharmacognosy wikipedia , lookup
Drug design wikipedia , lookup
Neuropsychopharmacology wikipedia , lookup
Drug discovery wikipedia , lookup
Psychedelic therapy wikipedia , lookup
Drug interaction wikipedia , lookup
Pharmacokinetics wikipedia , lookup
Theralizumab wikipedia , lookup
Discovery and development of direct thrombin inhibitors wikipedia , lookup
Transcript
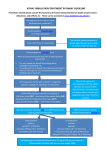
Pharmacogenetic Testing Prior to Initiation of Warfarin Therapy Introduction Warfarin’s anticoagulant effect is mediated through its ability to block hepatic synthesis of functional vitamin K-dependent clotting factors, specifically factors II, VII, IX, and X.2 Warfarin inhibits the intrahepatic vitamin K cycle and thus impairs vitamin K-dependent gamma-carboxylation.2 Recently, the gene encoding the vitamin K epoxide cycle has been identified (VKORC1).8 Alterations in the VKORC1 gene can affect the ability of warfarin to impair the activity of the vitamin K-dependent factors and, therefore, alter response to therapy.9 VKORC1 variants (polymorphisms) are thought to account for as much as 25% of the interindividual variation in required warfarin dose while about 10% of this variation is due to CYP2C9 variants.4,9 The VKORC1 genetic variant that is a marker for warfarin hypersensitivity is designated (-)1639 G>A.7,10 Carriers of this variant require significantly lower maintenance doses of warfarin than noncarriers.7 The frequency of (-)1639 G>A varies with ethnicity; it is more common in Chinese,10 less common in Caucasians, and even less common among those of African American ancestry.11 Variability in an individual’s response to a given drug may be due to a number of different genetic factors. Genetic variants may influence drug response by altering (1) drug metabolizing enzymes, (2) drug target(s) or receptor(s), and/or (3) drug transport protein(s) or a combination thereof.1 Pharmacogenetics is the investigation of the genetic basis of drug response. Unlike traditional therapeutic drug monitoring, a significant advantage of pharmacogenetic testing is that investigation can be undertaken prior to initiation of drug therapy to enable more effective initial dose stratification or identify specific situations in which a specific therapy will not be effective.1 Warfarin is an ideal drug target for the application of pharmacogenetic testing due to its narrow therapeutic index, large interindividual variation in dose requirements and high frequency of identified genetic variants that have an impact on both its effect and metabolism.2,3 Daily maintenance doses of warfarin in study subjects range widely, from 1.25 mg to 34 mg.4 Given this significant variation in required dose and warfarin’s narrow therapeutic index, the anticoagulant effect of warfarin must be regularly monitored. This is accomplished using a mathematical conversion of the prothrombin time (PT).2 The PT is converted to the International Normalized Ratio (INR) based on the mean PT of the population and responsiveness of the test reagent (INR = [patient PT/mean PT]ISI).2 The recommended therapeutic range for patients on warfarin therapy is an INR value between 2.0 and 3.0.2 Below this range, patients are at risk of thrombosis while supratherapeutic INR values increase the potential for bleeding.4–7 There is a sharp increase in bleeding risk when the INR is above the therapeutic range such that the risk of hemorrhage doubles with each one point rise in INR.5 Warfarin Metabolism Warfarin is metabolized in the liver through the cytochrome P450 (CYP) system.2 The CYP drug metabolizing enzyme 2C9 is responsible for the metabolism of the biologically active isomer of warfarin (S-warfarin), as well as other agents including certain antiepileptics (phenytoin), antidiabetic drugs (glipizide, tolbutamide), nonsteroidal anti-inflammatory agents and antihypertensive agents (losartan).1,12,13 Genetic variants in the CYP2C9 system can impair the degree to which warfarin is metabolized, thereby altering drug response.13 Common genetic variants (polymorphisms) of CYP2C9, designated CYP2C9*2 and CYP2C9*3, have been shown to decrease the in vivo clearance of warfarin therapy.14 In CYP2C9*2 homozygotes, metabolism of warfarin is reduced by 40% and in those who are homozygous for CYP2C9*3, warfarin metabolism is reduced by almost 90%.12,14 During initiation of warfarin therapy, individuals with the CYP2C9*2 and/or CYP2C9*3 genotype are more likely than noncarriers to have INR values greater than 4 and suffer a twofold to threefold elevated bleeding rate.14 Carriers of these variants, furthermore, require more frequent dose adjustments during warfarin initiation and take longer to reach a stable maintenance dose.15 The CYP2C9*2 and CYP2C9*3 polymorphisms are common, occurring in up to 30% of Caucasian populations, while both of these variants are less common in African and Asian populations.12 It has been estimated that 18% of Caucasians have both VKORC1 and one of the CYP2C9 variants16 and these individuals therefore require significantly lower doses of warfarin to achieve a therapeutic response and prevent overdose.4,10,16 Bleeding Complications Bleeding is the most clinically significant complication associated with warfarin therapy.4–6 Risk for bleeding is highest during warfarin initiation as required dose is not easily predicted in the individual patient.4–6 Commonly occurring genetic variants in warfarin’s target enzyme (or the enzyme system largely responsible for its metabolism) account for much of the observed variation in warfarin’s anticoagulant effect.7 Individuals who carry these common genetic variants require lower maintenance doses of warfarin.4,7 The application of pharmacogenetic testing prior to the start of warfarin therapy to identify these genetic variants, therefore, can identify a population that is hypersensitive to warfarin therapy, avoiding the potential for serious and life-threatening bleeding complications.3,4,7 1 Predicting warfarin dose at therapy initiation is enhanced by genotyping for CYP2C9*2 or *3 and (-)1639 G>A and consideration of other factors.4,7,10,15 For each CYP2C9*2 variant, required war farin dose is reduced by 19% and reduction in dose is 29% with CYP2C9*3 alleles.4 In determination of optimum warfarin dose, other factors such as patient age, intercurrent illness(es), nutrition, drug-drug interactions and body surface area should be taken into account.4,7 Certain drugs such as amiodorone, isoniazid, and phenylbutazone have been shown to decrease warfarin clearance and, therefore, reduce the required dose while rifampin and barbiturates have been shown to enhance warfarin metabolism, increasing the required amount of drug.17 Warfarin (P450 2C9 and VKORC1) . . . . . . . . . . . 511460 Synonyms DME Genotyping; Warfarin Genotyping Specimen Whole blood or LabCorp buccal swab kit (buccal swab collection kit contains instructions for use of a buccal swab.) Volume 7 mL whole blood or LabCorp buccal swab kit Minimum Volume 3 mL whole blood or two buccal swabs Container Lavender-top (EDTA) tube, yellow-top (ACD) tube, or LabCorp buccal swab kit Storage Instructions Maintain specimen ambient, or refrigerate at 4°C. Causes for Rejection Frozen specimen; hemolysis; quantity not sufficient for analysis; improper container; one buccal swab Use Warfarin, the coumarin derivative, is the most widely prescribed anticoagulant for thromboembolic disorders. This test detects variants within two genes, CYP2C9 and VKORC1, that influence the response to warfarin therapy. Understanding different rates of warfarin metabolism can help define optimal patient-specific warfarin dosage. Limitations The metabolism of drugs is also influenced by ethnicity, diet, and other medications. All factors should be considered prior to initiating new therapy. Methodology Polymerase chain reaction (PCR); gel electrophoresis Pharmacogenetic testing for warfarin de tects the presence of CYP2C9*2, *3, and VKCORC1 (-)1639 G>A variants. CYP2C9 DNA analysis is performed by allele-specific PCR followed by electrophoresis to detect the *2 and *3 alleles. VKORC1 DNA analysis is performed by PCR followed by restriction enzyme digestion to detect (-)1639 G>A. Individuals with one or more of these variant alleles are hypersensitive to warfarin therapy and have an increased risk for bleeding if warfarin therapy is initiated at standard doses. The dose of warfarin can be individually determined based on CYP2C9 and VKORC1 genotype, patient age, body surface area, and concurrent drug therapies.4 For the most current information regarding test options, including specimen requirements and CPT codes, please consult the online Test Menu at www.LabCorp.com. References 9. Rieder MJ, Reiner AP, Gage BF, et al. Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N Engl J Med. 2005 Jun 2; 352(22):2285-2293. 10. Yuan HY, Chen J-J, Lee MTM, et al. A novel functional VKORC1 promoter polymorphism is associated with inter-individual and inter-ethnic differences in warfarin sensitivity. Hum Mol Genet. 2005; 14(13):1745-1751. 11. Geisen C, Watzka M, Sittinger K, et al. VKORC1 haplotypes and their impact on the inter-individual and inter-ethnical variability of oral anticoagulation. Thromb Haemost. 2005; 94:773-779. 12. Adcock DM, Koftan C, Crisan D, Kiechle FL. Effect of polymorphism in the cytochrome P450 CYP2C9 gene on warfarin metabolism. Arch Pathol Lab Med. 2004 Dec; 128:1360-1363. 13. Takahashi H, Echizen H. Pharmacogenetics of warfarin elimination and its clinical implications. Clin Pharmacokinet. 2001; 40(8):587-602. 14. Voora D, Eby C, Linder MW, et al. Prospective dosing of warfarin on cytochrome P-450 2C9 genotype. Thromb Haemost. 2005; 93:700-705. 15. Higashi MK, Veenstra DL, Kondo LM, et al. Association between CYP2C9 genetic variants and anticoagulation-related outcomes during warfarin therapy. JAMA. 2002 Apr 3; 287(13):1690-1698. 16. Marsh S, King CR, Porche-Sorbet RM, Scott-Horton TJ, Eby CS. Population variation in VKORC1 haplotype structure. J Thromb Haemost. 2006; 4:473-474. 17. Wells PS, Holbrook AM, Crowther NR, Hirsh J. Interactions of warfarin with drugs and food. Ann Intern Med. 1994 Nov 1; 121(9):676-683. 1. Ensom MH, Chang TK, Patel P. Pharmacogenetics: The therapeutic drug monitoring of the future? Clin Pharmacokinet. 2001; 40(11);783-802. 2. Hirsh J, Dalen JE, Anderson DR, et al. Oral anticoagulants: Mechanism of action, clinical effectiveness, and optimal therapeutic range. Chest. 2001 Jan; 119(1):8S-21S. 3. Bodin L, Verstuyft C, Tregouet D-A, et al. Cytochrome P450 2C9 (CYP2C9) and vitamin K epoxide reductase (VKORC1) genotypes as determinants of acenocoumarol sensitivity. Blood. 2005 Jul 1; 106(1):135-139. 4. Gage BF, Eby C, Milligan PE, Banet GA, Duncan JR, McLeod HL. Use of pharmacogenetics and clinical factors to predict the maintenance dose of warfarin. Thromb Haemost. 2004; 91:87-94. 5. Landefeld CS, Rosenblatt MW, Goldman L. Major bleeding in outpatients treated with warfarin: Incidence and prediction by factors known at the start of outpatient therapy. Am J Med. 1989 Aug; 87(2):153-159. 6. White RH, Beyth RJ, Zhou H, Romano PS. Major bleeding after hospitalization for deep-venous thrombosis. Am J Med. 1999 Nov; 107(5):414-424. 7. Sconce EA, Khan TI, Wynne HA, et al. The impact of CYP2C9 and VKORC1 genetic polymorphism and patient characteristics upon warfarin dose requirements: Proposal for a new dosing regimen. Blood. 2005 Oct 1; 106(7):2329-2333. 8. Li T, Chang C-Y, Jin D-Y, Lin P-J, Khvorova A, Stafford DW. Identification of the gene for vitamin K epoxide reductase. Nature. 2004 Feb 5; 427:541-544. 2 ©2014 Laboratory Corporation of America® Holdings All Rights Reserved L4627-0914-4