* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Identification and Localization of Carbon Concentrating
Mycoplasma laboratorium wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Gene prediction wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Transformation (genetics) wikipedia , lookup
Gene expression profiling wikipedia , lookup
Designer baby wikipedia , lookup
Genetic engineering wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
Genetically modified crops wikipedia , lookup
Community fingerprinting wikipedia , lookup
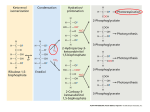
Identification and Localization of Carbon Concentrating Components in Chlamydomonas reinhardtii INTRODUCTION • • Most crops, including wheat, rice and soybean use C 3 photosynthesis C4 and CAM photosynthesis overcome the tendency of RuBisCO to wastefully fix oxygen rather than carbon dioxide (photorespiration) while C3 does not KEY QUESTIONS RESULTS • PIPELINE EFFICIENCY • • What proteins are essential to the functioning of the CCM in Chlamydomonas? Which proteins should be transformed into C3 plants to improve photosynthetic efficiency? How can we optimize the High Throughput Tagging Pipeline? PCR 67% Cloning 95% METHODOLOGY http://www.citruscollege.edu/lc/archive/biology CHLAMYDOMONAS REINHARDTII • Photosynthetic apparatus similar to land plants • Unicellular • Heterotrophic and autotrophic photosynthetic apparatus • Partially sequenced genome http://protist.i.hosei.ac.jp CARBON CONCENTRATING MECHANISM • CCM increases the concentration of carbon dioxide available to the initial carboxylase of the Calvin cycle, RuBisCO • Benefits include an increased tolerance to low concentrations of inorganic carbon, reduced photorespiration and a greater tolerance to water stress • Multiple genome saturated mutant screen • Touchdown PCR • Products were run through Gel Electrophoresis Identification and Amplification Maurino, Veronica G., et al., 2010 • Synthetic detours naturally occurring in cyanobacteria were installed in Arabidopsis thaliana, to bypass photorespiration McGrath, Justin M., et al., 2014 • Models predict the benefits of engineering a full cyanobacterial CCM into a C3 leaf • Increased: o Leaf photosynthesis by 25% o Soybean yield by 15% o Water use efficiency by 20% REFERENCES Eckardt, Nancy A. "Gene Regulatory Networks of the Carbon-Concentrating Mechanism in Chlamydomonas reinhardtii." The Plant Cell Online 24.5 (2012): 1713-1713. Fang, Wei, et al. "Transcriptome-wide changes in Chlamydomonas reinhardtii gene expression regulated by carbon dioxide and the CO2-concentrating mechanism regulator CIA5/CCM1." The Plant Cell Online 24.5 (2012): 1876-1893. Hom, Erik FY, and Andrew W. Murray. "Niche engineering demonstrates a latent capacity for fungal-algal mutualism." Science 345.6192 (2014): 94-98 Kebeish, Rashad, et al. "Chloroplastic photorespiratory bypass increases photosynthesis and biomass production in Arabidopsis thaliana." Nature biotechnology 25.5 (2007): 593-599. Maurino, Veronica G., and Christoph Peterhansel. "Photorespiration: current status and approaches for metabolic engineering." Current opinion in plant biology 13.3 (2010): 248-255. Wang, Yingham, and Deqiang Duanmu. "Carbon dioxide concentrating mechanism in Chlamydomonas reinhardtii: inorganic carbon transport and CO2 recapture." pringer Science and Business Media (2010): n. pag. Print. 95% • Purified products were run against samples of known concentration to determine DNA concentration Purification • • Localization 65% ELECTROPORATION EFFICIENCY 467 DNA fragments were successfully amplified 83 fully amplified genes were transformed into E.coli 52 genes were localized efficiently LOCALIZATION • PCR PRODUCTS • • Cre04.g229300_F, a suggested RuBisCO Activase coding protein The concentrated green pocket indicated that the gene was localized in the pyrenoid In total, 83 genes were localized, and 60% were identifiable • Gel purified genes were cloned in frame with yellow fluorescent protein, CrVenus • Ampicillin resistance • Transformed E.coli by heat shock Gibson Assembly Image provided courtesy of Dr. Luke Mackinder Transformation of CCM into C3 crops could increase crop yield REVIEW OF LITERATURE Transformation • PCR ALTERATIONS • Plasmids were extracted then cut with the restriction enzyme Eco-RV to confirm successful cloning Plasmid Sequencing • Cassettes were incubated in a 16oC water bath for 5 minutes • DNA was added, and a shock was administered at 800V and 25 uF Electroporation and Plating 6% DMSO CONCLUSIONS • • Transformation plates were screened for strong Venus expressing colonies using the Typhoon Imaging System • Confocal imaging determined subcellular localization Localization DISCUSSION • • • • • • 4.5% DMSO • • High Throughput Tagging Pipeline can be successfully utilized to amplify and localize putative photosynthetic genes of the CCM in the alga Chlamydomonas reinhardtii Electroporation efficiency is negatively affected by increased time transformation cassettes were left in a 16oC water bath Failed PCR primers can be recovered by increasing DMSO levels and decreasing extension times 82 genes will be imaged and analyzed at a later date Complementation vectors will be constructed with different resistance markers Increased availability of information to other labs will assist in all realms of C.reinhardtii research It is estimated that there will be about 11 billion people living on the earth by 2100 New innovations need to be made to increase food production Transforming a viable CCM into crop plants augments photosynthetic productivity, increasing crop yield www.pflanzenforschung.de