* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download lecture notes-enzyme-web
Multi-state modeling of biomolecules wikipedia , lookup
Nicotinamide adenine dinucleotide wikipedia , lookup
Cooperative binding wikipedia , lookup
Inositol-trisphosphate 3-kinase wikipedia , lookup
Restriction enzyme wikipedia , lookup
Beta-lactamase wikipedia , lookup
Transferase wikipedia , lookup
Alcohol dehydrogenase wikipedia , lookup
Lactoylglutathione lyase wikipedia , lookup
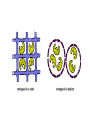
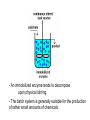
Models for More Complex Enzyme Kinetics • Allosteric enzymes - Some enzymes have more than one substrate binding site. - Allostery or cooperative binding: the binding of one substrate to the enzyme facilitates binding of other substrate molecules. Models for More Complex Enzyme Kinetics The rate expression in this case is Vm [ S ]n d[S ] v dt K m "[ S ]n n is cooperativity coefficient; n>1 indicates positive cooperativity; n can be determined by rearranging the above equation as v '' ln n ln[S ] ln K m Vm v v And plotting ln V v m versus ln[S]. Models for More Complex Enzyme Kinetics • Insoluble substrate wood chips, cellulosic residues -Access to the active site on these biopolymers by enzyme is limited by enzyme diffusion. -The number of reaction sites exceeds the number of enzyme molecules. -This is opposite that of the typical situation with soluble substrates, where access to the enzyme’s active site by substrate limits reaction. Models for More Complex Enzyme Kinetics • Insoluble substrate If considering - the initial product formation rate and - the reaction is first order in terms of the enzyme-substrate complex concentration, yields, v V max, S [ E ] K eq [ E ] Vmax,S k2[S0 ] K eq k des / k ads Factors Affecting Enzyme Kinetics • pH effects - on enzymes - enzymes have ionic groups on their active sites. - Variation of pH changes the ionic form of the active sites. - pH changes the three-Dimensional structure of enzyme. - on substrate - some substrates contain ionic groups - pH affects the ionic form of substrate affects the affinity of the substrate to the enzyme Factors Affecting Enzyme Kinetics • Temperature - on the rate enzyme catalyzed reaction d[ P] v k 2 [ ES ] dt k2=A*exp(-Ea/R*T) T k2 v - enzyme denaturation T d[ E ] kd [E] Denaturation rate: dt kd=Ad*exp(-Ea/R*T) kd: enzyme denaturation rate constant; Ea: deactivation energy Immobilized Enzyme Systems • Enzyme immobilization: To restrict enzyme mobility in a fixed space. Advantages: - Easy separation from reaction mixture, providing the ability to control reaction times and minimize the enzymes lost in the product. - Re-use of enzymes for many reaction cycles, lowering the total production cost of enzyme mediated reactions. - Ability of enzymes to provide pure products. - Possible provision of a better environment for enzyme activity - Study of the action of membrane-bound intracellular enzyme Immobilized Enzyme Systems • Methods of Enzyme Immobilization: - Entrapment - Surface immobilization - Cross-linking Immobilized Enzyme Systems Entrapment immobilization is based on the localization of an enzyme within the lattice of a polymer matrix or membrane. - to retain enzyme - allow the penetration of substrate. It can be classified into matrix and micro capsule types. Immobilized Enzyme Systems Entrapment - matrix entrapment - membrane entrapment (microencapsulation) Immobilized Enzyme Systems - matrix entrapment Matrix materials: organics: polysaccharides, proteins, carbon, vinyl and allyl polymers, and polyamides. e.g. Ca-alginate, agar, K-carrageenin, collagen Enzyme + polymer solution → polymerization → extrusion/shape the particles inorganics: activated carbon, porous ceramic and diatomaceous earth. Shapes: - particle - membrane - fiber Immobilized Enzyme Systems - membrane entrapment - Regular semipermeable membrane: nylon, cellulose, polysulfone and polyacrylate. - Microencapsulation: Microscopic hollow sphere are formed. The sphere contain the enzyme solution and is enclosed within a porous membrane. Immobilized Enzyme Systems Entrapment challenges: - enzyme leakage into solution - diffusional limitation - reduced enzyme activity and stability - lack of control micro-environmental conditions. It could be improved by modifying matrix or membrane. Immobilized Enzyme Systems Surface immobilization According to the binding mode of the enzyme, this method can be further sub-classified into: - Physical Adsorption - Ionic Binding - Covalent Binding Immobilized Enzyme Systems Surface immobilization Physical Adsorption is based on the physical adsorption of enzyme protein on the surface of water-insoluble carriers. - If a suitable carrier is found, this method can be both simple and cheap. - The active sites and activity of enzymes are less affected. - Desorption of enzyme takes place because of weak attraction. - Non-specific of other protein or substances will affect the properties of enzyme. Immobilized Enzyme Systems Physical Adsorption: Weak forces: Van der Waals or dispersion Materials: Inorganic: almumina, silica, porous glass, ceramics. Organic: e.g. cellulose, starch, activated carbon. Carbon nano-tube (Kim, Jeong Yun, Special Publication - Royal Society of Chemistry, 2004) Immobilized Enzyme Systems Ionic binding: Interaction forces: ionic bonds. Features: similar to that of physical adsorption. Polysaccharides and synthetic polymers having ion-exchange centers are usually used as carriers. Immobilized Enzyme Systems Covalently binding is the formation of covalent bonds between the enzyme and the support matrix. Interaction forces: covalent bonds. Features: - may alter the conformational structure and active center of the enzyme, resulting in major loss of activity and/or changes of the substrate. - the binding force between enzyme and carrier is so strong that no leakage of the enzymes occurs. Immobilized Enzyme Systems Covalent binding: To select the type of reaction for enzyme covalent immobilization: - do not cause loss of enzymatic activity. - the active site of the enzyme must be unaffected by the reagents used. Immobilized Enzyme Systems Covalent binding: The functional groups on the supports that may take part in this binding are listed below: e.g. amino group, carboxyl group, sulfhydryl group, hydroxyl group, phenolic group etc. Immobilized Enzyme Systems Cross-linking: to cross link enzyme molecules with each other using agents such as glutaraldehyde. Features: similar to covalent binding. Several methods are combined. Summary of Immobilization Methods Methods of Enzyme immobilization: - Entrapment - matrix - membrane (microencapsulation) - Surface immobilization - physical adsorption - ionic binding - covalent binding - Cross-linking Immobilized Enzyme Reactors Recycle packed column reactor: - allow the reactor to operate at high fluid velocities. - a substrate that cannot be completely processed on a single pass Fluidized Bed Reactor: - a high viscosity substrate solution - a gaseous substrate or product in a continuous reaction system - care must be taken to avoid the destruction and decomposition of immobilized enzymes - An immobilized enzyme tends to decompose upon physical stirring. - The batch system is generally suitable for the production of rather small amounts of chemicals.