* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
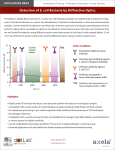
Download Figure S1. Architecture of genetic elements in bacteria different of K
Genetic drift wikipedia , lookup
Behavioural genetics wikipedia , lookup
Human genetic variation wikipedia , lookup
Heritability of IQ wikipedia , lookup
Population genetics wikipedia , lookup
Pathogenomics wikipedia , lookup
Genetic testing wikipedia , lookup
Expanded genetic code wikipedia , lookup
Microevolution wikipedia , lookup
Public health genomics wikipedia , lookup
Genome (book) wikipedia , lookup
History of genetic engineering wikipedia , lookup
Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is © The Royal Society of Chemistry 2017 FigureS1.ArchitectureofgeneticelementsinbacteriadifferentofK-12MG1655 WeperformedthesameanalysisasforE.colistrainK-12MG1655inE.colistrainsK-12 W3110 and BL21 (DE3), in Salmonella typhimurium SL1344, and in Pseudomonas aeruginosastrainsPA14andPAO1(see,TableS2andS3).A)ConsensusarchitectureofE. coli K12 MG1655, B) Summary of the consensus architecture of all the other bacteria, where genetic elements in colors are those conserved as in K-12 MG1655 and those in grayhavelimitedinformationtogetconsensus.Weidentifiedthatthegeneticelements conserved in all bacteria are: transcription start site, translation start codon, translation stop codon, ribosomal binding sites, genes overlap in 4 bp and the average distance betweentheTSSandtheRBSrangefrom20to40bp.