* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download HLA-G - DTU CBS
Survey
Document related concepts
Transcript
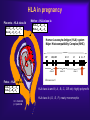
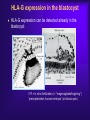
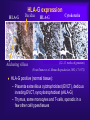
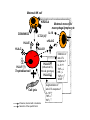
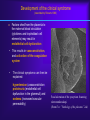
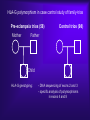
Reproductive Immunology The HLA System in Pregnancy ’Why did your mother not reject you?’ Thomas Hviid, MD, PhD Dept. of Clinical Biochemistry Roskilde University Hospital Denmark Topics Introduction: The semi-allogenic fetus Fundamental aspects of MHC/HLA and HLA class Ib – ’Classical’ vs ’non-classical’ MHC/HLA genes – Gene structure, polymorphisms, expression, alternative splicing, functions etc Certain complications of pregnancy in relation to HLA/HLA-G – (Recurrent) spontaneous abortions, IVF, pre-eclampsia Pregnancy and HLA diversity – Reproductive selection mechanisms (’mating preferences’) and HLA The ’semi-allogenic fetus’ Medawar & Billingham, Nature, 1953 Four hypotheses: – The conceptus lacks immunogenicity – Significant lowering of the immune response during pregnancy – The uterus is an immunoprivileged site – Immune barrier elaborated by the placenta: • Tolerance to the semi-allognic fetus by the maternal immune system seems mainly an active mechanism: – Fetal tissue prevented from being recognized as foreign tissue and/or being rejected by the maternal immune system Hypotheses/concepts to explain maternal tolerance of the fetus HLA/HLA-G expression by the trophoblast The Th1/Th2 balance Regulatory CD4+CD25+ T cells Others – – – – – – – – Leukemia inhibitory factor (LIF) Indoleamine 2,3-dioxygenase (IDO) Suppressor macrophages Hormones CD95 and its ligand Annexin II Lowered complement activity Hidden trophoblast antigens (Thellin et al, review 2000) HLA and the ’semi-allogenic fetus’ Human Leucocyte Antigen (HLA) Major Histocompatibility Complex (MHC) Several classes of HLA genes: – HLA class Ia (classical HLA class I antigens; on almost all cells) • HLA-A, HLA-B, HLA-C – HLA class Ib (non-classical HLA class I antigens) • HLA-E, HLA-G, HLA-F – HLA class II (expressed on antigen presenting cells, B cells) • HLA-DR, HLA-DP, HLA-DQ Discovered / rejection of transplants (Jean Dausset 1952/1953) The function of HLA / MHC was elucidated in the 1970s (Zinkernagel & Doherty 1974) MHC/HLA molecules present antigen peptides to T cells via the T cell receptor Antigen peptides (eg pathogenes) are recognized in combination with an individual’s own variant of HLA The classical HLA class Ia molecules are highly polymorphic HLA-A10 HLA-B12 HLA-Cw5 HLA-A3 HLA-B5 HLA-Cw7 HLA-A23 HLA-B12 HLA-Cw1 HLA-A11 HLA-B16 HLA-Cw8 HLA-A25 HLA-B40 HLA-Cw2 HLA-A26 HLA-B8 HLA-Cw5 HLA-A2 HLA-B27 HLA-Cw6 HLA-A28 HLA-B17 HLA-Cw5 HLA-A19 HLA-B14 HLA-Cw8 HLA-A19 HLA-B15 HLA-Cw2 HLA-A25 HLA-B12 HLA-Cw1 HLA-A24 HLA-B8 HLA-Cw4 The non-classical HLA class Ib molecules are nearly monomorphic HLA-G HLA-E HLA-F HLA-G HLA-E HLA-F HLA-G HLA-E HLA-F HLA-G HLA-E HLA-F HLA-G HLA-E HLA-F HLA-G HLA-E HLA-F HLA-G HLA-E HLA-F HLA-G HLA-E HLA-F HLA-G HLA-E HLA-F HLA-G HLA-E HLA-F HLA-G HLA-E HLA-F HLA-G HLA-E HLA-F HLA in pregnancy Placenta – HLA class Ib HLA-Gm, -Em, -Fm, -Cm HLA-Gp, -Ep, -Fp, -Cp Mother – HLA class Ia HLA-Am, -Bm, -Cm HLA-Am, -Bm, -Cm Human Leucocyte Antigen (HLA) system Major Histocompatibility Complex (MHC) DP DQ DR class II Fetus – HLA class Ia HLA-Am, -Bm, -Cm HLA-Ap, -Bp, -Cp m = maternal p = paternal B C class III E A G F class I Chromosome 6 HLA class Ia and II (-A, -B, -C, -DR etc): highly polymorfic HLA class Ib (-G, -E, -F): nearly monomorphic HLA-G expression in the blastocyst HLA-G expression can be detected already in the blastocyst IVF = in vitro fertilization (= ”reagensglasbefrugtning”) ’preimplantation human embryos’ (or blastocysts) HLA-G expression in the blastocyst/embryo Detection of HLA-G mRNA in around 40% of preimplantation human embryos (Jurisicova et al 1996, Cao et al 1999) No detection of HLA-G mRNA in human embryos (but only 11 embryos investigated) (Hiby et al 1999) Detection of soluble HLA-G in some human embryo culture supernatants from IVF after 46-72 hrs (in total >1000 embryo cultures) (Fuzzi et al 2002, Sher et al 2004, Noci et al 2005, Yie et al 2005) 36% sHLA-G pos. of single embryo cultures (Noci et al 2005) No detection of sHLA-G in human embryo cultures (Lierop et al 2002) Expression of HLA-G mRNA and sHLA-G has been associated with an increased cleavage rate, as compared to embryos lacking HLA-G (Jurisicova et al 1996, Yie et al 2005) Soluble HLA-G and success of IVF The pregnancy rate in women who have embryos transferred from cultures where sHLA-G is detected is significantly higher than that in women who have only embryos transferred from sHLA-G negative cultures (Fuzzi et al 2002, Sher et al 2004, Noci et al 2005, Yie et al 2005) Pregnancy and live births are observed in sHLA-G- neg. IVF cycles; however, the rate of spontaneous abortions is higher in the HLA-G-negative group (25%) vs. the HLA-G-positive group (11%) (Yie et al 2005) HLA-G expression HLA-G Decidua HLA-G Cytokeratin (12.-13. weeks of gestation) Anchoring villous (From Emmer et al. Human Reproduction 2002; 17:1072) HLA-G positive (normal tissue): – Placenta extravillous cytotrophoblast (EVCT), dedicua invading EVCT, syncytiotrophoblast (sHLA-G) – Thymus, some monocytes and T-cells, sporadic in a few other cell types/tissues HLA-G gene, mRNA, protein, isoforms 1-domain 2-domain -2m 3-domain HLA-G1 Other alternatively spliced HLA-G mRNA isoforms exist +14 bp: 45 % 14 bp deleted: 55 % 14 bp del polymorphism (Harrison et al 1993) HLA-G alleles (DNA sequences) HLA-G 5’URR/Promotor*) Exon 2 alleles -725 -201 31 35 54 57 69 93 100 107 110 130 188 236 241 258 nt 3741 G*010101 C or G G ACG CGG CAG CCG GCC CAC GGC GGA CTC CTG CAC GCA TTC ACG - G*010102 C A --- --- --- --A --- --T --- --- --- --- --- --- --- --- +14 bp G*010103 C A --- --- --- --A --- --- --- --T --- --- --- --- -C- --- +14 bp G*010104 C G --- --- --- --- --T --- --- --- --- --- n.d. n.d. n.d. n.d. n.d. G*010105 n.d. n.d. --- --- --- --- --- --- --- --T --- --- --- --- --- --- - G*010106 n.d. n.d. --- --- --- --- --- --- --- --- --- --- --T --- --- --- n.d. G*010107 n.d. n.d. --- --- --- --A --- --T --- --T --- --- --- --- --- --- n.d. G*010108 C G --- --- --- --A --- --- --- --- --- --- --- --- --- --- - G*0102 n.d. n.d. --- --- -G- --- --- --- --- --- --- --- --- --C --- --- n.d. G*0103 C (or T) G T-- --- --- --- --- --- --- --- --- --- --T n.d. n.d. --- +14 bp G*010401 C A --- --- --- --A --- --- --- --- A-- --- --- --- --- --- - G*010402 n.d. n.d. --- --- --- --C --- --- --- --- A-- --- --T --- --- --- n.d. G*010403 n.d. n.d. --- --- --- --- --- --- --- --- A-- --- --- --- --- --- - G*0105N C A --- --- --- --A --- --T --- --- --- TG --- n.d. n.d. --- +14 bp G*0106 C A --- --- --- --A --- --T --- --- --- --- --- --- --- -T- +14 bp G*0101g**) n.d. n.d. --- --A --- --- --- --- --- --- --- --- --- n.d. n.d. --- +14 bp G*G3d5**) n.d. --- --- --- --A --- --T --T --- --- --- --- n.d. n.d. --- +14 bp n.d. Exon 3 3’UTR*) Exon 4 Amino acid substitution HLA-G polymorphisms Consensus: Only a handful of HLA-G alleles with amino acid substitutions Around 15 HLA-G alleles at the DNA level (Bodmer et al. Hum Immunol 1999; 60:361) Threonin Serin Deletion of a cytosine (codon 129/130) frameshift Leucin Isoleucin HLA-A HLA-G (After Ober & Aldrich, J Reprod Immunol 1997; 36:1-21 and Parham, Eur J Immunogenetics 1992; 19:347-359. Based on work by Bjorkman et al, Nature 1987; 329:512-518). HLA-G*0105N HLA-G*0105N is a so-called null allele – One base pair is deleted in exon 3 of the HLA-G gene (Most likely) non-functional HLA-G1 and HLA-G5 (full membrane and soluble isoforms) Clinical data on HLA-G*0105N homozygotes shows that HLA-G1 and –G5 are not essential for fetal survival However, normal HLA-G2 – G4 and G6/G7 are encoded and these isoforms seem to be functional in much the same way as G1/G5 (Sala et al 2004, Le Discorde et al 2005) HLA-G functions Possible contributions of HLA-G in the implantation process: 1) Attachment of the blastocyst to the endometrium • HLA-G has been found to be involved in cellular adhesion (Ødum et al 1991) 2) Trophoblast invasion of uterine tissue and maternal spiral arteries • HLA-G is expressed by endovascular trophoblast cells and may be a modulator of angiogenesis (Le Bouteiller et al) 3) Trophoblast interaction with maternal immune effector cells • HLA-G interacts with receptors on immune cells Allo-cytotoxic T lymphocyte (CTL) response Augmentation of the allo-CTL response stimulator cell responder T cell T cell receptor HLADR4 IL-10 TNF- INF- HLADR1 Inhibition of allo-CTL response HLA-DR4 HLA-G (Kapasi et al 2000) HLA-G IL-10 TNF- INF- RECURRENT MISCARRIAGE AND PREUNCOMPLICATED PREGNANCY ECLAMPSIA Upregulation of the Th1 response, downregulation of Th2: IL-2 INF- (TNF-) Th2 cytokine production: IL-4 IL-5 IL-10 IL-13 The Th1/Th2 balance HLA-G/sHLA-G??? Successful pregnancy more often correlated with a Th2type response than Th1 However, the Th1/Th2 concept may be too simplistic Functions of HLA-G Several in vitro studies have shown that HLA-G and HLA-E protect against Natural Killer-mediated cell lysis Functions of HLA-G Suppression of allo-reactive cytotoxic T cells ‘Mixed Lymphocyte Reaction’ (MLR) stimulator cell responder T cell CD4+ responder T cell T cell receptor HLADR4 Secretion of soluble HLA-G5 HLADR1 HLA-DR4 inhibitory receptor (ILT-2, p49 ?) HLA-G1 K562 Inhibition of T cell allo-proliferation (Carosella et al. Immunol Today 1999; 20:60 / Riteau et al. J Reprod Immunol 1999; 43:203) (Lila et al PNAS 2001; 98:12150) Maternal NK cell KIR2DL4 CD94/NKG2 ILT-2/(-4)*) sHLA-G HLA-E HLA-C Maternal monocyte/ macrophage/lymphocyte IL-10 ? HLA-G1 HLA-F (?) Trophoblast cell FETUS Cell lysis Influence, interact with or modulate Secretion of the specific factor HLA-G (influenced by HLA-G genotype) HLA-G Inhibition of allo-CTL response ? IL-10 ?? TNF- INF- TGF-1 VEGF Augmentation of allo-CTL response ? IL-10 ?? TNF- INF- Summary/ Acceptance of the semi-allogenic fetus… No expression of polymorfic HLA class Ia and II on fetal trophoblast cells in the placenta Placenta – HLA class Ib HLA-Gm, -Em, -Fm , -Cm HLA-Gp , -E p, -Fp , -Cp NB! Natural Killer cell-mediated lysis Expression of non-polymorfic HLA class Ib molecules by trophoblast: HLA-G (and HLA-E and –F) Fetus – HLA class Ia HLA-Am, -Bm, -Cm HLA-Ap , -B p, -Cp This expression profile may influence the cytokine profile in favour of maintaining pregnancy m = maternal p = paternal Mother – HLA class Ia HLA-Am, -Bm, -Cm HLA-Am, -Bm, -Cm Regulatory T cells (Tregs) CD4+CD25bright(FoxP3+) – CD4: co-receptor binds to MHC class II – CD25: alpha-chain of IL-2 receptor – FoxP3: transcription factor essential for differentiation into CD4+CD25+ Tregs (Sasaki et al 2004) Tregs important for their potential to prevent autoimmune diseases May also play important roles in tolerance induction in organ transplantations Regulatory T cells in reproduction Mice: – Transfer of CD4+CD25+ Tregs from normal pregnant mice to abortion-prone mice prevented spontaneous abortion – Decidual TGF-beta and LIF were upregulated in Treg-treated mice (Zenclussen et al 2006) Humans: – Pregnancy is associated with an increase in circulating CD4+CD25+ Tregs, and also an increase in decidua, during early pregnancy (Somerset et al 2004; Tilburgs et al 2006) – Proportion of decidual CD4+CD25bright Tregs has been shown to be significantly lower in cases of spontaneous abortion compared to induced abortion – Decreased CD4+CD25+ Tregs in spontaneous abortion might induce maternal lymphocyte activation to the semi-allogenic fetus (Sasaki et al 2004) HLA and certain complications of pregnancy… Recurrent spontaneous abortions = Recurrent miscarriage Early pregnancy loss NB Chromosomal abnormalities are the most frequent cause of spontaneous abortions – however, many are ’unexplained’ and some may be due to immunological dysfunction HLA and recurrent miscarriage (RM) Prospective studies in inbred populations clearly show an influence of HLA genes or closely linked loci on reproductive processes, (studies in the Hutterites by Ober and co-workers) Many studies have focused on a possible increased sharing of HLA alleles/haplotypes between the mother and the father(/the fetus) in RM. However, ’HLA sharing’ is a controversial issue and lacks evidence. Specific HLA-DR alleles are associated with increased risk of RM – Meta-analysis (18 published/unpublished case-control studies): HLA-DRB1*01 risk factor (OR 1.3; 95%CI 1.1-1.6) (Christiansen et al 1999) – HLA-DRB1*03 risk factor in patients with 4 or more miscarriages and a significantly increasing trend with increasing number of previous miscarriages (OR 1.4; 95%CI 1.1-1.9) (Kruse et al 2004) Soluble HLA-G assays (plasma/serum) Some confusion exists regarding the detection of sHLA-G in blood samples It seems that sHLA-G can be detected in all plasma samples from pregnant and non-pregnant women, while sHLA-G can only be detected in some serum samples from at least non-pregnant women (and from men) Low amounts of sHLA-G may be ’trapped’ in the clot formation in serum samples. Therefore, the serum sHLA-G levels may be lower than the plasma sHLAG level, and blood with low amounts of sHLA-G might be sHLA-G negative when serum samples are investigated Levels of sHLA-G in maternal blood (plasma) Maternal sHLA-G levels do not change substantially during a normal course of pregnancy Soluble HLA-G levels of non-pregnant and pregnant women seems to be very similar Therefore, a substantial part of the sHLA-G detected in maternal circulation may be produced by immunocompetent cells of the mother Reduced levels of sHLA-G in maternal plasma may be associated with pre-eclampsia, spontaneous abortion and placental abruption (sHLA-G < 9.95 ng/ml RR 7.1; 3 trim) (Steinborn et al 2003) Pregnancy after IVF and soluble HLA-G In 20 women who experienced an early spontaneous abortion, the preovulatory sHLA-G conc. was significant reduced compared to women with an intact pregnancy. The same difference was observed during monitorering of sHLA-G levels in intact pregnancy vs early spontaneous abortion until 9th week of gestation (p < 0.0001). (Pfeiffer et al 2000) HLA-G genetics and women with RM Study (listed by type and chronology) HLA-G polymorphism Penzes et al. (1999) Type of study Technical comments Casecontrol PCR-RFLP of 21 RSA exons 2 couples and 3; low resolution of alleles Yamashita et al. (1999) Casecontrol Pfeiffer et al. (2001) Casecontrol PCR-SSCP of exons 2, 3 and 4. DNA sequencing of intron 4 DNA sequencing of exons 2 and 3 Aldrich et al. (2001) Ober et al. (2003) Cohort PCR-SSOP, exons 2 and 3 Cases 20 RSA couples 78 RSA women (3 abortions; 28% secondary aborters) 113 couples (3 abortions; one live born child) Controls Results 72 healthy, unrelated individuals Negative. However, a trend towards a higher frequency of the G*01012 allele in the controls 54 healthy fertile Negative; no controls (27 implication of females/27 polymorphism in males) intron 4 52 women G*010103 and G*0105N associated (1 successful with RSA pregnancies) Cohort (15 years) DNA sequencing of 42 Hutterite 5’URR; 18 SNPs non-RSA women Hviid et al. (2002, 2004a) Casecontrol DNA sequencing of exons 2 and 3, polymorphisms in exons 4 and 8 61 RSA couples (3 abortions; 38% secondary aborters) 47 fertile couples/93 fertile women (2 normal pregnancies) Tripathi et al. (2004) Casecontrol Casecontrol 120 RSA women (3 abortions; primary aborters) 120 fertile women (3 live births) Abbas et al. (2004) ACLA/lupus anticoagulant positive RSA women? 300 included; 180 lost? PCR-SSOP analysis of exons 2 and 3; typing of G*0106? G*0104 or G*0105N in either partner associated with increased risk for abortion Increased risk for abortion in couples both carrying a –725G allele (OR 2.7; 95%CI 1.1-7.1) +14/+14 bp HLA-G genotype of female associated with RSA (OR 2.7; 95%CI 1.16.5). More RSA women carried the G*0106 allele (15%) compared to controls (2%) (n.s.) -14/+14 bp HLA-G genotype of female associated with RSA Higher frequency of G*010103 in RSA women (n.s.) Negative Negative G*010103 and G*0105N G*0104 and G*0105N -725G in 5’URR +14/+14-bp genotype Trend for G*0106 -14/+14-bp genotype Trend for G*010103 HLA-G 14-bp genotypes in in vitro fertilisation (IVF) - a pilot study (Hviid et al 2004) Association of the 14-bp HLA-G polymorphism to the outcome of IVF treatments ? Two groups of couples attending IVF: – ”Uncomplicated” pregnancy with twins after first IVF treatment (n = 15) – 3 IVF treatments without pregnancy/implantation (n = 14) HLA-G genotyping Clinical and laboratory data / eg. embryo grade, inseminated oocytes etc 14-bp HLA-G genotype of women in in vitro fertilization (IVF) treatments or with recurrent miscarriage Mantel-Haenszel statistics (combined 2x2 tables) : P < 0.01 HLA-G and RM: Odds ratio 2.7 [95% CI 1.1-6.5] (Hviid et al 2004) Membrane-bound and soluble HLA-G mRNA levels in relation to the 14 bp sequence polymorphism in trophoblast cells Membrane-bound HLA-G Soluble HLA-G Comparison of HLA-G1/G2/G3 (-14 bp) and HLA-G1/G2/G3 (+14 bp) mRNA levels in heterozygous samples 70 p = 0.0156 40 p = 0.0313 35 S9 S10 30 S1 25 S4 20 S7 S3 60 Relative Fluorescence Units (RFU) Relative Fluorescence Units (RFU) Comparison of HLA-G5/G6 (-14 bp) and HLA-G5/G6 (+14 bp) mRNA levels in heterozygous samples 15 10 S8 50 40 S3 30 S7 20 10 5 S1 S8S10 S4 0 0 HLA-G RT-P CR product 545 bp HLA-G1/G2/G3 (-14 bp) mRNA HLA-G RT-P CR product 559 bp HLA-G1/G2/G3 (+ 14 bp) m RNA HLA-G RT-P CR product 417 bp HLA-G5/G6 (-14 bp) mRNA HLA-G RT-P CR product 431 bp HLA-G5/G6 (+14 bp) mRNA (Hviid et al 2003) HLA-G alleles / alternative splicing Genomic DNA -14 bp HLA-G*010101 +14 bp 3’UTR polymorphism HLA-G*010102 HLA-G*010103 G1 (-92 bp) G5/G6 (-92 bp) mRNA isoforms G2(/G4) (-92 bp) G1 G2/G4 G3 G5 G6 G1 (+14 bp) G2/G4 (+14 bp) G3 (+14 bp) G5 (+14 bp) G6 (+14 bp) G1 (+14 bp) G2/G4 (+14 bp) G3 (+14 bp) G5 (+14 bp) G6 (+14 bp) (Hviid et al 2003, Rousseau et al 2003) HLA-G / alleles / mRNA Conclusions… HLA-G alleles are associated with different HLA-G mRNA isoform expression profiles The HLA-G mRNAs including the 14 bp sequence in exon 8 are processed further than HLA-G mRNAs with the sequence deleted. This may influence HLA-G mRNA stability Soluble HLA-G in serum and the HLA-G genotype Italian serum samples Danish serum samples All samples *) HLA-G genotype Total HLA-G5/sG1 detected Total HLA-G5/sG1 detected Total HLA-G5/sG1 detected 14/14 55 12 23 5 78 17 14/+14 66 11 48 13 114 24 +14/+14 28 0 14 0 42 0 Total 149 23 85 18 234 41 2 test for observed distribution of serum samples with HLA-G5/sHLA-G1 detected in relation to HLA-G genotype and the expected independent distribution according to the overall HLA-G genotype frequencies/proportions (14/14: 13.7; 14/+14: 20.0; +14/+14: 7.4): 2 = 9.04; df = 2; P = 0.011 *) (Hviid et al 2004, Rizzo et al 2005) Soluble HLA-G levels in plasma Associations of soluble HLA-G (sHLA-G) plasma levels and HLA-G alleles For example, in four healthy individuals: In comparison to HLA-G*01011: – ”Low secretors”: – ”High secretors”: G*01013 and G*0105N G*0104 (Rebmann/van der Ven and co-workers 2001) Functional significance HLA-G gene sequence variation influences individual HLA-G expression Low or aberrant expression of membrane-bound and soluble HLA-G may have implications for NK-cell and T-cell interactions and cytokine profiles during pregnancy And hence - may influence the outcome of the pregnancy….. HLA and certain complications of pregnancy… Pre-eclampsia and HLA-G (pre-eclampsia = ”svangerskabsforgiftning”) Pre-eclampsia - ’a disease of theories’ Second half of pregnancy: – hypertension – proteinuria – (oedema) 2-7% of all pregnancies World-wide still a prominent cause of maternal and fetal mortality The fetus may also be compromised – Intrauterine growth retardation, low birth-wight, prematurity, and intrauterine asphyxia The etiology involves probably a combination of genetic and environmental risk factors Pre-eclampsia – patogenesis ? The presence of a placenta is both necessary and sufficient to cause the disorder. A fetus is not required as pre-eclampsia can occur with hydatidiform mole (Chun et al 1964) • Pre-eclampsia may develop with abdominal pregnancy (Piering et al 1993) • Central to management, is delivery, which removes the causative organ, the placenta. Placental pathoanatomy / pre-eclampsia (From Khong et al. British Journal of Obstetrics and Gynaecology 1986; 93:1049-1059) Step one (1. and 2. trimester?) Step two (3. trimester) (From Rubin & Farber, ”Pathology”; 1988) Development of the clinical syndrome (described by Roberts 1989) Factors shed from the placenta to the maternal blood circulation (cytokines and trophoblast cell elements) may result in endothelial cell dysfunction • This results in vasoconstriction, and activation of the coagulation system • The clinical symptoms can then be explained: hypertension (vasoconstriction), proteinuria (endothelial cell dysfunction in the glomeruli) and oedema (increased vascular permeability) Focal ulceration of the syncytium. Scanning electronmikroskopi. (From Fox: ”Pathology of the placenta” 2ed) Large epidemiological study concluded, that both the mother and the fetus contribute to the development of pre-eclampsia, and the fetus’ contribution is under influence of paternal genes (Lie et al 1998) Studies of genotypes in family trios mother-father-offspring Pre-eclampsia and HLA-G A role for HLA-G ? An obvious candidate gene Pre-eclampsia might be a consequence of an immunological maladaptation of the pregnant woman to the semi-allogenic fetus HLA-G and pre-eclampsia Several studies have found an aberrant or absent HLA-G expression in pre-eclamptic placentas both at the mRNA and protein level compared to control placentae – Colbern et al 1994 (mRNA) NB! Inconclusive – Hara et al 1996 – Troeger et al 1999 [abstract] – Lim et al 1997 – Goldman-Wohl et al 2000 (immunohistochemistry) (immunohistochemistry) (in vitro trophoblast cultures mRNA og protein) (in situ hybridisation) – O’Brien et al 2001 (mRNA and polymorphisms) – Association to +14 / +14 HLA-G genotypes – Yie et al 2004 (serum and placental HLA-G) HLA-G polymorphism in case control study of family-trios Pre-eclampsia trios (58) Mother Control trios (98) Father Child HLA-G genotyping: - DNA sequencing of exons 2 and 3 - specific analysis of polymorphisms in exons 4 and 8 Hypotheses The aberrant HLA-G expression in pre-eclampsia might be influenced by HLA-G genetics A higher number of +14/+14 exon 8 HLA-G genotypes in the pre-eclamptic offspring compared to the control group? (O’Brien et al 2001) HLA-G histo-incompatibility between mother and fetus? Frequencies of the 14 bp deletion polymorphism in exon 8 of the HLA-G gene in primipara family triads with a history of preeclampsia and controls Mothera NO. (%) PREECLAMPSIA Fatherb NO. (%) Offspringc NO. (%) Mothera NO. (%) CONTROLS Fatherb NO. (%) Offspringc NO. (%) Allele: 14 bp +14 bp n= 44 36 80 55.0 45.0 41 39 80 51.3 48.8 38 42 80 47.5 52.5 86 54 140 61.4 38.6 92 48 140 65.7 34.3 96 44 140 68.6 31.4 Genotype: 14 bp/14 bp 14 bp/+14 b +14 bp/+14 bp n= 12 20 8 40 30.0 50.0 20.0 10 21 9 40 25.0 52.5 22.5 10 18 12 40 25.0 45.0 30.0 29 28 13 70 41.4 40.0 18.6 30 32 8 70 42.9 45.7 11.4 31 34 5 70 44.3 48.6 7.1 a Alleles: P = 0.393; Fisher’s exact test. Genotypes: P = 0.472; 2 = 1.50, df 2. b Alleles: P = 0.045; Fisher’s exact test. Genotypes: P = 0.106; 2 = 4.49, df 2. c Alleles: P = 0.003; Fisher’s exact test. Genotypes: P = 0.004; 2 = 11.21, df 2. The +14 bp/+14 bp genotype vs others: P = 0.002; Fisher’s exact test. Odds ratio = 5.57 [95% CI 1.79-17.31]. (Hylenius et al 2004) HLA-G expression in pre-eclamptic placentas GeneChip data on HLA-G mRNA expression: Control_A 1517,42 Control_B 969,89 Control_C 445,3 baseline mean 974,69 PE_A 598,46 PE_B 744,79 PE_C 556,99 experiment mean 632,91 fold change -1,54 lower bound of FC upper bound of FC -0,72 -2,5 HLA-G mRNA expression reduced in pre-eclamptic placentas (Hviid et al 2004) Conclusions / HLA-G and pre-eclampsia The HLA-G genotype of the fetus and the HLA-G expression in the placenta seems to be involved in the pathogenesis of preeclampsia Subfecundity correlates with pre-eclampsia… HLA-G and organ transplantation Expression of soluble HLA-G in serum and HLA-G by heart and liver/kidney grafts have been associated with significant better prognosis and fewer rejection episodes – (…remember…only in some serum samples can HLA-G be detected; and only some grafts are positive for HLA-G expression) (Lila et al 2002, Creput et al 2003) HLA-G and organ transplantation Recent independent study / heart transplants: – Two groups of heart transplant patients (n=9 and n=10) • One group displayed a significant increase (p>0.001) in sHLA-G during the first month after transplantation (>50 ng/ml) • The other group maintained low levels of sHLA-G (<30 ng/ml) – 50% of the patients with low levels of sHLA-G had recurrent severe rejection episodes, whereas patients with high levels of sHLA-G did not have any episodes of recurrent severe rejection (Luque et al 2006;Hum Immunol 67,257) HLA-G and organ transplatation Ongoing study / Transplantation Unit, Rigshospitalet, Cph: – Monitoring sHLA-G with different assays, HLA-G genetics and regulatory T cells in lung and heart transplantations After all there might be some parallels in mechanisms inducing tolerance during pregnancy and during organ transplantation sHLA-G might have potential as a therapeutic agent in transplantation MHC class Ib in other species……. Rat – Transcripts for a soluble form of the RT1-E MHC class Ib molecule have been detected in placenta; could have a regulatory role on NK-cell function (Solier et al 2001) Non-human primates – Rhesus monkey: expression in placenta of a Mamu-AG gene with a soluble isoform (Ryan et al 2002) – Baboon: placental expression of soluble/membrane-bound PaanAG MHC class Ib proteins (Langat et al 2002) MHC class Ib in other species……. Mouse: Qa-2 antigen / Preimplantation embryo development (Ped) gene phenotype Present on oocytes/blastocysts Major influence on preimplantation embryonic cleavage rate and division (Warner et al 1993, Exley & Warner 1999) Presence of the Ped gene phenotype correlates to heavier birth weight and larger litters (Warner et al 1991) During development there is a selection in favour of presence of the Ped gene phenotype in the offspring (Exley & Warner 1999) MHC/HLA and mating preferences HLA / mating preferences / body odors T-shirts experiments…………………. Studies of inbred mouse strains and humans have revealed avoidance of mates with a very high number of matching MHC alleles. (In humans, female students dislike the smell of T-shirts worn by males with identical HLA alleles…) (e.g. Potts et al 1991, Wedekind et al 1995, Ober et al 1997) MHC homozygote deficiency reported in small, isolated populations (e.g. Ober et al) In human populations the frequency of MHC heterozygotes are typically higher than expected by chance HLA / mating preferences / body odors Possible mechanisms….. – Avoids inbreeding (e.g. Bateson 1983) – MHC heterozygosity confers a selective advantage against multiple-strain infections (Penn et al 2002) – In mice, the MHC seems to influence fertilization and pregnancy outcome; maybe even oocyte/sperm selection (e.g. Wedekind et al 1996, Rühlicke et al 1998, Exley & Warner 1999). In the pub…… Can I buy you a drink? Sorry – I don’t think you’re my type…. Can I buy you a drink? Buzz off – I think you’re my (HLA) type…. HLA / mating preferences / body odors Detection of MHC-mediated body odor may result from the close linkage between the MHC loci and olfactory receptor genes (Fan et al 1996, Amadou et al 1999) MHC/HLA-specific odors may be soluble MHC proteins, odor molecules bound selectively to MHC proteins, or by-products of MHC-specific bacteria colonization in skin or axillae (Yamazaki et al 1999, Vincent & Revillard 1979, Pearse-Pratt et al 1999) (Ehlers et al 2000. Genome research 10:1968-1978) Summary MHC/HLA in reproduction Mating preferences seem to be influenced by MHC/HLA diversity Fertilization Weak evidence for MHC/HLA-mediated effects on spermatogenesis Heterozygote advantage Heterozygotes at the MHC/ HLA loci may provide a broader immune response Early embryo development and implantation HLA-G expression associated with cleavage rate and implantation success Maternal genome Paternal genome Balance between foetal/paternal and maternal interests? Some HLA-G/MHC polymorphisms may work in favour of the foetus, others in favour of maternal interests? Foetal growth and survival Some evidence that HLA haplotypes and HLA-G polymorphism are associated with birth weight, risk of abortion and immuneadaptation Deficiency of MHC/HLA homozygotes in isolated populations: frequency of MHC heterozygotes in human populations higher than expected Collaborators Anne-Marie Nybo Andersen Alessandra Balboni Olavio R. Baricordi Ole B. Christiansen Maria Teresa Grappa Sine Hylenius Anne Mette Høgh Christina Kruse Anette Lindhard Mads Melbye Loredana Melchiorri Nils Milman Lone G. Nielsen Marina Stignani Roberta Rizzo Christina Rørbye