* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Construction of the Heart`s Conduction Tree via Prim`s Algorithm
Quantium Medical Cardiac Output wikipedia , lookup
Coronary artery disease wikipedia , lookup
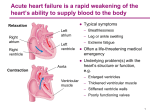
Heart failure wikipedia , lookup
Mitral insufficiency wikipedia , lookup
Lutembacher's syndrome wikipedia , lookup
Myocardial infarction wikipedia , lookup
Electrocardiography wikipedia , lookup
Dextro-Transposition of the great arteries wikipedia , lookup
Congenital heart defect wikipedia , lookup
Arrhythmogenic right ventricular dysplasia wikipedia , lookup
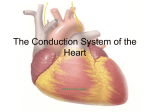
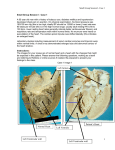
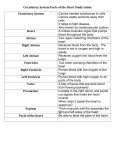
POSTER 2016, PRAGUE MAY 24 1 Construction of the Heart’s Conduction Tree via Prim’s Algorithm Leonie KORN Chair of Medical Information Technology, Helmholtz-Institute for Biomedical Engineering, RWTH Aachen University, 52074 Aachen, Germany [email protected] Abstract. The heart’s excitation is a complex process which is often modelled for several cases of applications. Realtime capability is often postulated but not guaranteed due to the huge amount of used cells. Therefore, in this paper an abstract conduction system was developed to reduce complexity. To distinguish between different cardiac regions, a segmentation of the heart has been performed. The cardiac conduction system is divided in its regions of tissue in which the prim’s algorithm has been applied to identify their directed minimal spanning trees for conduction orientation. As a result an abstract anatomic model which includes the orientation of the main direction of propagation has been developed. The model’s potential impact is to reduce complexity for real-time modelling of the heart’s excitation. Keywords Heart excitation, Tree, Prim’s algorithm, Modelling Real-time systems. prim’s algorithm is given. Section 3 shows the results of modelling. The paper ends with a discussion. 2. Conduction tree construction 2.1. Modelling the heart’s anatomy The heart’s anatomy has been modelled using a Computer Tomographical (CT) image series of 76 images with a resolution of 512x512 (from [2]). A pre-segmentation of this dataset has been performed by windowing. This was done according to the Hounsfield scale which describes quantitatively the radiodensity in gray-scale values. It is defined by the linear attenuation coefficient of water μwater as seen in Eqn. 1. μtissue − μwater (1) HU = 1000 ∗ μwater Typical values for different substances can be seen in Tab. 1. Thus, the heart is separated from surrounding pulmonary Substance 1. Introduction The heart’s pumping function is triggered by electric impulses. These impulses have their origin at the sinoatrial node and spread over the atrium to the atrioventricular node, the left and right bundle branches, through the purkinje fibres into the ventricle’s myocardium [1]. This initiates the essential contraction of the ventricles, so that blood is pumped into the cardiovascular system. More closely these impulses are cardiac action potentials, characteristically shaped and generated at every heart cell. The excitation in the heart cells is directly related to the recorded biosignals in and on the body volume conductor. The large amount of heart cells result in a huge complexity regarding biosignal’s modelling but are significant for the signal’s shapes. That’s why for real-time applications an abstraction of this complex conduction system is necessary. Therefore, in this paper an abstract model of the heart’s conduction system has been developed. This paper is organized as follows. In section 2 the construction of the conduction tree is described. In detail, modelling of the heart’s anatomy and the description of the modified Hounsfield sclae [HU] Air -1000 -500 Lung Fat -100 to -50 0 Water 15 Cerebro-spinal fluid 30 Kidney Blood 35 to 45 Muscle 10 to 45 Liver Soft Tissue Bone 40 to 60 100 to 300 700 to 3000 Tab. 1. Typical values of the Hounsfield scale [4]. tissue, bones and air. For well perfused soft tissue the Hounsfield scale is not sufficient, since the transitions between various soft tissue is not clear. Therefore, a manual segmentation with ITK-SNAP was done to distinguish between atrium, ventricles, endocardium and epicardium of the heart (see Fig. 3). With this tool the Sagittal plane, the Coronal plane and the Transversal plane are visible anytime. 2 L. KORN, CONSTRUCTION OF THE HEART’S CONDUCTION TREE VIA PRIM’S ALGORITHM CT-Dataset approx. 40 mio voxel ITK-SNAP: Segmentation Ventricle selection Surface data approx. 40.000 vertices MeshLab: Smoothing Decimation Surface data approx. 400 vertices Fig. 1. Steps of the heart segmentation. z[mm] Orientation in these three orthogonal planes is simple because of a shared cursor. The manual segmentation is done due to the atlas of human anatomical cross sections [3]. To generate a convenient abstraction of the heart’s conduction system, the surface data from the segmented parts from the heart was processed with MeshLab. The heart’s surface data was smoothed with a HC Laplacian Smooth filter and decimated by a Quadric Edge Collapse Decimator. Here, the heart’s surface consists of triangulations. The separation between endocardium and epicardium as well as the separation of the left and right ventricle was realised. This is useful for further processing. Within the segmentation process the CTdataset with approx. 40 million voxels has been reduced to approx. 400 vertices to create a basis for the heart’s conduction tree (see Fig. 1). In Fig. 2 the result of the heart’s segmentation, as well as x[mm] y[mm] Fig. 2. Segmented ventricle surface. the reduced number of vertices on the heart’s surface can be seen for the left and right ventricle. 2.2. Modified prim’s algorithm A tree structure was used to model the spatial propagation of the heart’s excitation. Thus, a predefined conduction orientation is desired to reduce complexity, so that realtime capability can be achieved. The conduction system of the heart includes different tissue areas. In these areas the shape and the conduction velocity of the cell’s action potentials differ from each other. In Tab. 2 the different conduction velocities of the heart areas are given. These differences are important since they are directly related to the shape of Heart area Sinoatrial node Myocradium atrium Atrioventricular node Conduction velocity in m/s < 0.01 1.0 - 1.2 0.02 - 0.05 Bundle of His 1.2 - 2.0 Bundle branches 2.0 - 4.0 Purkinje fibres 2.0 - 4.0 Ventricle myocardium 0.3 - 1.0 Tab. 2. Conduction velocities of different heart areas [4]. biosignals measured in the body volume conductor or on the body volume conductor’s surface. Therefore, the heart is divided into the following tissue areas: sinoatrial node, left and right atrium, the atrioventricular node, the bundle of His, left and right bundle branches and the left and right ventricle’s myocardium. The vertices of the conduction tree were found manually for the sinoatrial node, left and right atrium, the atrioventricular node, the bundle of His and the left and right bundle branches. In contrast the vertices of the left and right ventricle’s myocardium were found automatically using a modified prim’s algorithm. The algorithm of prim generates the minimal spanning tree for a weighted, undirected graph. This minimal spanning tree includes all vertices, so that the total weight of all edges is minimized. The graph G = (V, E) contains the amount of vertices V and the amount of edges E. In E all available connections between different vertices V are enclosed. Every edge (u, v) ∈ E is dedicated a weight w(u, v). Finding the minimal spanning tree is an iterative process. Initially the undirected graph G contains all vertices V . In a first step, searching for the edge with a minimum weight to the root r is done and the corresponding vertex is included as a new vertex into the directed graph A. Within every next step the lightest edge from a vertex within the vertices from the directed graph A to one of the vertices from the undirected graph G is determined and added. Only this way it is ensured that, in the end, graph A involves every vertex. A pseudo code of the algorithm’s functionality is shown in Algo. 1 [5]: POSTER 2016, PRAGUE MAY 24 3 Fig. 3. 3D-view of the segmented heart. G: Graph w: weights r: root V: Vertices Q: Queue of priority π(u): predecessor of u Adj(u): list of adjacent vertices from u dist(u): distance from u to spanning tree they have the shortest distance to the end points of the right and left bundle branches. In the next step the modified prim’s algorithm can be applied, starting at the chosen roots. First only vertices on the inner surface, the endocardium, are considered for the prim’s algorithm and are included into the directed graph A. Then the vertices on the epicardium, the outer surface of the heart, are added to the graph according to the method of prim’s algorithm. All manual and automatic found minimal spanning trees were merged to get a predefined orientation of the heart’s conduction tree. Algorithm 1 Prim’s algorithm(G,w,r) Q ← V [G] for all u ∈ Q do dist[u] ← ∞ π[u] ← 0 dist[r] ← 0 end for while Q = 0 do u ← FindMin[Q] for all v ∈ Adj[u] do if v ∈ Q and w(u, v) < dist[v] then π[v] ← u dist[u] ← w(u, v) end if end for end while return A Initializing 3. Results As a result the abstract anatomy of the heart’s conduction system modelled via the modified prim’s algorithm is shown in Fig. 4. The different spanning trees of the tissue areas are represented in various colours. The direction of the -140 -160 Atrium Tawara Ventricle 1, right Ventricle 2,right Ventricle 3,right Ventricle posterior,left Ventricle anterior,left -180 In this special case, the prim’s algorithm has been modified for finding the heart’s conduction tree. Therefore, the distances between all vertices of the heart’s surface data are determined and saved in an array of weights. A connection of two vertices is only added as an edge into the array of edges E, with its determined weight, if the distance stays under a certain threshold. Only by this it’s guaranteed that the concave inner surface of the ventricles can be depicted correctly. The right ventricle has three propagation strands starting near the apex. The left ventricle has two propagation strands, one posterior and one anterior passing. Thus, in the right ventricle there are three independent and in the left ventricle two independent minimal spanning trees implemented. The roots of those minimal spanning trees are chosen, so that z [mm] -200 -220 -240 260 240 40 220 20 200 0 180 -20 -40 160 y [mm] -60 140 -80 120 x [mm] -100 Fig. 4. The heart’s conduction tree. complete conduction tree can be followed from the atrium to the bundle branches into the left and right myocardium. 4 L. KORN, CONSTRUCTION OF THE HEART’S CONDUCTION TREE VIA PRIM’S ALGORITHM 4. Conclusion The results show an abstraction of the heart’s conduction system in a way that a predefined propagation direction exists. The characteristic anatomy of the heart is well recognizable. The spread of excitation in the heart has a hierarchical structure. Therefore, choosing a tree structure to model the spatial expansion of the impulses seems reasonable. The reason to use a modified prim’s algorithm in this model is founded by the physiological timing of the impulses’ propagation. First the excitation spreads from the sinoatrial node, to the atrium into the left and right bundle branches. Initially in the ventricle’s myocardium the excitation starts on the inner surface of the heart, the endocardium. The main direction of propagation points from the apex into the direction of the sinoatrial node. Afterwards the excitation transits to the outer surface, the epicardium, so that the whole ventricle’s myocardium is activated. This characteristic spread of excitation can be ensured by using the modified prim’s algorithm. For future work the developed conduction system can be used to model spatial distribution of action potentials in the heart. All vertices are representing different heart cells and the edges can be weighted with area characteristic conduction velocities. With its tree structure the developed model is well-suited especially for using cellular automata [6]. Further processing of this spatial distribution of action potentials can result in real-time modelling of biosignals. Acknowledgements The research presented in this paper benefited from the input of Daniel Rüschen, M.Sc. and Univ.-Prof. Dr.-Ing. Dr. med. Steffen Leonhardt, RWTH Aachen University, who supervised this work. References [1] FOGOROS, R.N. Electrophysiological Testing. John Wiley & Sons, 2012. [2] OsiriX. DICOM sampla image set MAGIX. http://www.osirixviewer.com/datasets/MAGIX.zip. [3] MÖLLER, T.B., REIF, E. Taschenatlas der Schnittbildanatomie: Thorax, Herz, Abdomen, Becken. Thieme, 2011. [4] IAIZZO, P. Handbook of Cardiac Anatomy, Physiology and Devices. Humana Press, 2009. [5] CORMEN, T.H., LEISERSON, C.E., RIVEST, R.L., STEIN, C. Minimum Spanning Trees in Introduction To Algorithms. MIT Press, 2001. [6] YE, P., ENTCHEVA, E., GROSU, R., SMOLKA, S.A. Efficient modeling of excitable cells using hybrid automata. Proc. of CMSB, 2005, vol. 2, p. 216 - 227. About Authors. . . Leonie KORN was born in Aachen, Germany, on May 3, 1990. She received the M.Sc. degree in Electrical Engineering with specialisation on Biomedical Engineering from the RWTH Aachen University, Germany, in October 2015. Currently she is working towards her doctor degree at the Philips Chair of Medical Information Technology, RWTH Aachen University. Her research interests include physiological modelling and biosignal processing with a focus on ventricular assist devices.